Figure 3.
Genes and hallmark pathways distinguish late-phase maturation fate
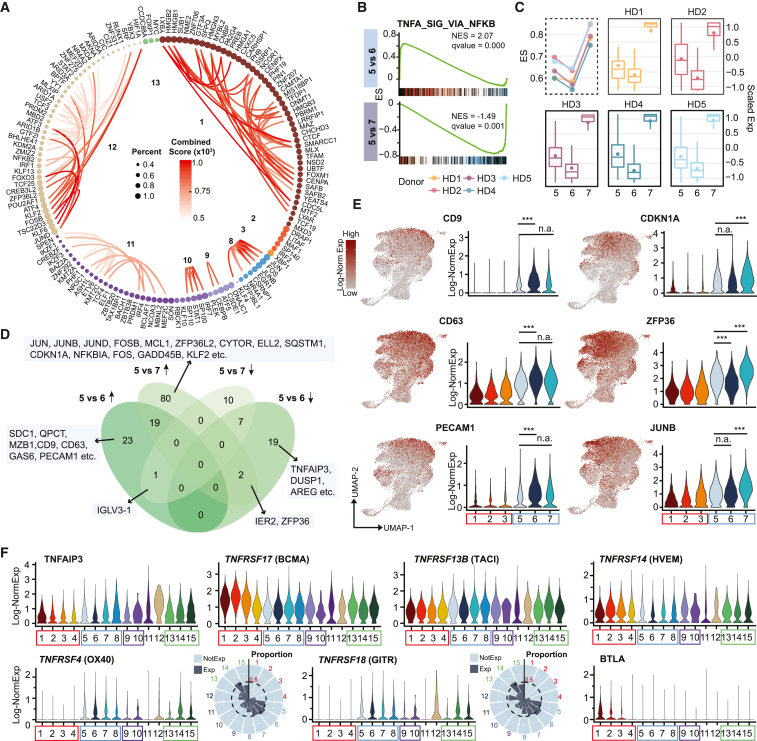
(A) Cell-cluster-associated TFs; each node represents a TF, colored by associated cell cluster and scaled by the expressed percentage of cells in the cluster. The lines between nodes are inferred protein-protein interactions from the STRING database. The redder the line, the more confident the inferred interactions. Numbers in the circles show the assigned cell cluster ID.
(B) GSEA of the most indicated pathways. TNFα signaling via NF-κB that are differentially enriched between 5 vs. 6 and 5 vs. 7.
(C) TNFα signaling via NF-κB hallmark pathway ES separated for each of the five individual subjects (in dashed box and y axis is on the left) and the distribution of scaled corresponding pathway enriched maturation-associated DEG expression in c5, c6, and c7 (in solid boxes and y axis is on the right).
(D) Comparison of top DEGs between c5 vs. c6 and c5 vs. c7 (see star methods), based on the sign of average log fold-change (avglogFC), the DEGs were divided into up-/downregulated in c6 or c7 groups. Venn diagram shows the results of the DEG comparison.
(E) Gene expression examples from the results of the comparisons in (D).
(F) Gene expression of genes from TNF family. The x axis is cell cluster ID and y axis is the log-normalized gene expression. Circular bar plot shows the proportion of cells in each cell subgroup showing expression of corresponding genes. Black dashed line indicates the proportion of 0.5 and numbers indicate the cell cluster IDs. NotExp, not expressed; Exp, expressed. ∗∗∗Bonferroni-adjusted p value <0.001; n.a., not available.