Figure 4.
Trophectoderm cells derived from iPSCs with risk alleles of rs917172 and rs12386620 show increased sensitivity to ZIKV infection
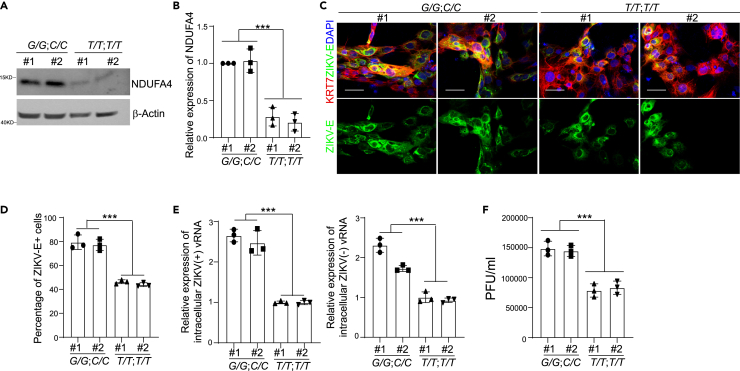
(A and B) Western blotting analysis (A) and quantification (B) of NDUFA4 protein expression in trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles. β-Actin was used as a loading control.
(C and D) Representative confocal images (C) and the quantification (D) of ZIKV-E staining in KRT7+ trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(E) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands in trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1).
(F) Viral titers of ZIKV in the supernatant of trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1) quantified by plaque assay. Data are representative of at least three independent experiments. p values were calculated by two-way ANOVA analysis; ∗∗∗p < 0.001.
See also Figure S4.