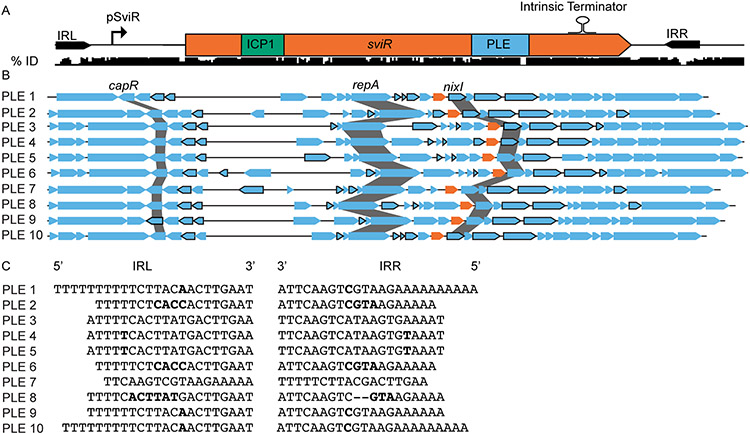
Figure 6). SviR is a core regulatory component of all PLEs discovered to date.
(A) SviR and its flanking repeats sequences are conserved across all ten PLEs. A nucleotide alignment of SviR and the flanking regions (represented by the black bars below the gene graph) shows the percent identity of the region across all ten PLEs. The ICP1 and PLE seed regions are highlighted in green and blue, respectively. The inverted repeats flanking SviR are marked by black arrows and the relative location of the SviR promoter and intrinsic terminator are annotated above the sequence, as determined by RACE and bioinformatic predictions. IRL = inverted repeat left, IRR = inverted repeat right.
(B) SviR is predicted to be a regulatory feature of all ten PLEs. Each of the ten PLEs is shown by a representative gene graph with each SviR allele in orange. ORFs outlined in black are predicted by IntaRNA to interact with the SviR PLE seed region with a ΔG° < −10.
(C) The inverted repeats flanking SviR from each of the ten PLEs. The mismatched bases between repeat sequences are highlighted in bold, and gaps in the repeat sequences are represented by hyphens. Despite the highly variable sequences between the 10 PLEs, the presence of inverted repeats is consistent across all PLEs.