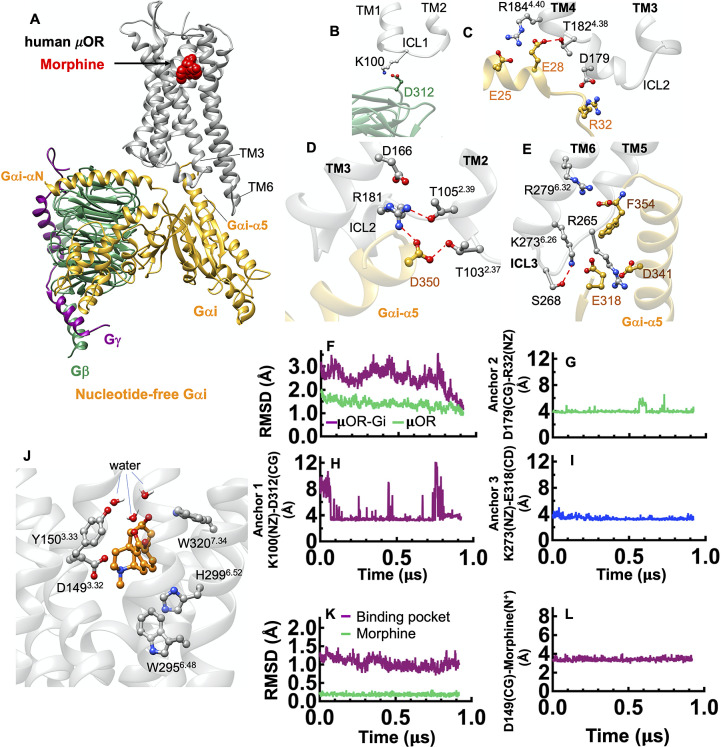
Fig. 3.
Gi protein binds the human μOR by forming ionic anchors to ICL1, ICL2 and the cytoplasmic end of TM6. (A) Structure of the human μOR–Gi protein complex derived from a ~950 ns MD simulation using Amber14. (B) The ionic anchor from the Gβ subunit to the ICL1. (C) Salt bridge anchors from the Gαi subunit to ICL2 and to the cytoplasmic end of TM4. (D) The network of polar interactions between ICL2 and the Gαi-α5 helix and (E) ionic anchors from the Gαi subunit to the ICL3 and the cytosolic end of TM6. (F) RMSD variation of the complex with time. Here, the RMSD calculated for the backbone atoms of the whole structures over the simulation and compared to the final snapshot. (G-I) Variation of the salt bridge anchors between Gi protein-μOR with time. The dotted red lines indicate hydrogen bonding. (J) Human μOR binding pocket after ~950 ns of MD simulation. The salt bridge between D149(CG) and morphine (the protonated N atom), locks morphine in the orthosteric binding pocket. (K) RMSD variation for the binding pocket and morphine with time. (L) The key salt bridge interaction between D1493.32 and the morphine protonated N atom (the protonated N atom) that holds morphine in tight contact with the human μOR.