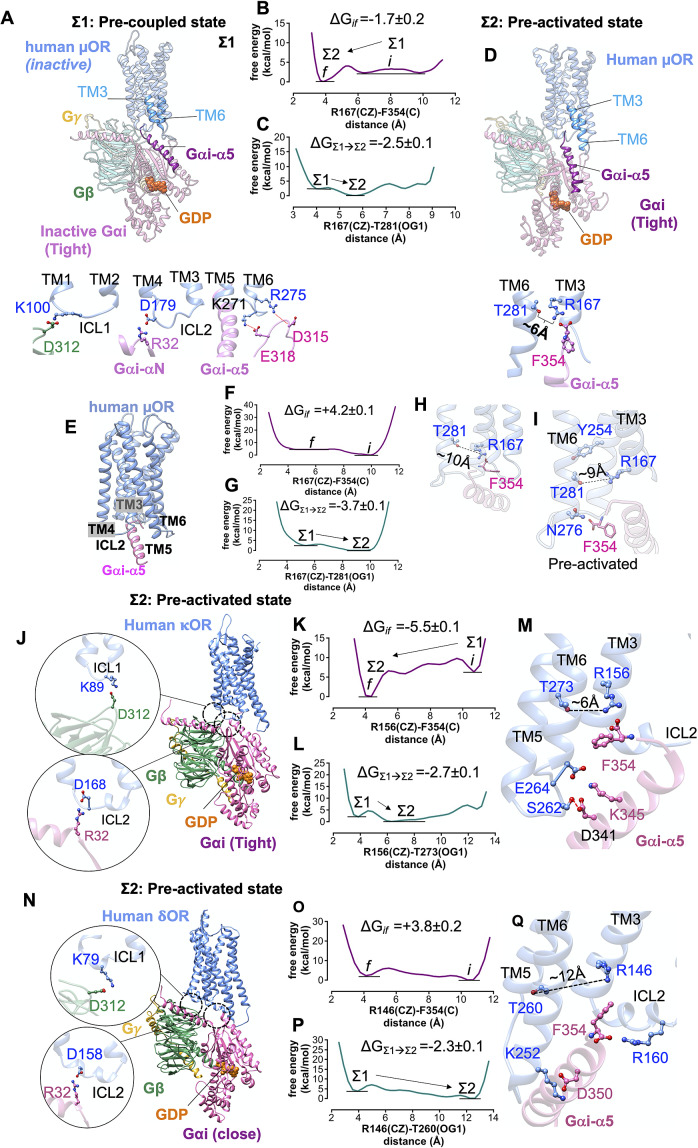
Fig. 5.
Formation of pre-activated complex (Σ2) between Gi protein and opioid receptors prior to ligand binding. (A) The structure of the pre-coupled state (Σ1 state), comprising the inactive human μOR and inactive Gi protein-bound GDP (tight); the Gi protein binds to the inactive μOR by forming salt bridge anchors to ICL1, ICL2 and ICL3. (B) MetaMD free energy profile for the salt bridge between the F354 (C) and R1673.50 (CZ). (C) MetaMD free energy for opening the polar interaction between R1673.50 (CZ) and T2816.34 (OG1) while F354 makes a strong salt bridge with R1673.50. (D) The structure of the pre-activated state (Σ2) showing open μOR (broken polar interaction R1673.50-T2816.34), and the salt bridge between F354-R1673.50. (E) The pre-activated complex (Σ2) between human μOR and the Gαi-α5 peptide (the rest of the Gi protein is eliminated) showing that formation of the pre-activated state is not dependent on a specific rigid-body orientation modelled in our Σ1 state. (F) MetaMD free energy profile for the interaction between the F354 (C) terminal carboxylate and R1673.50 (CZ). (G) MetaMD free energy for breaking the polar interaction between R1673.50 and T2816.34. (H) The salt bridge between F354 (C) and R1673.50 breaks the polar interaction between R1673.50-T2816.34. (I) Detailed structural analysis for the intracellular region of pre-activated complex between human μOR and the Gαi-α5 peptide. (J,N) The pre-activated complex between Gi protein and κOR/δOR, respectively. (K,O) MetaMD free energy profiles for the interaction between the F354 (C) and κOR-R1563.50 (CZ) and δOR-R1463.50 (CZ), respectively. (L,P) MetaMD free energy for breaking the coupling between TM3 and TM6 κOR: R1563.50-T2736.34 δOR: R1463.50-T2606.34. (M,Q) Detailed structural analysis for the intracellular region of pre-activated complex between human κOR/δOR and Gi protein. The weighted averages and the standard deviations were calculated for the converged period between the initial configuration before metaMD ‘i’ and the final conformation ‘f’ after metaMD calculations (Supplementary Figs S4–S6).