ABSTRACT
Antimicrobial acroautophagy/autophagy plays a vital role in degrading intracellular pathogens or microbial molecules in host-microbe interactions. However, microbes evolved various mechanisms to hijack or modulate autophagy to escape elimination. Vector-transmitted phloem-limited bacteria, Candidatus Liberibacter (Ca. Liberibacter) species, cause Huanglongbing (HLB), one of the most catastrophic citrus diseases worldwide, yet contributions of autophagy to HLB disease proliferation remain poorly defined. Here, we report the identification of a virulence effector in “Ca. Liberibacter asiaticus” (Las), SDE3, which is highly conserved among the “Ca. Liberibacter”. SDE3 expression not only promotes the disease development of HLB and canker in sweet orange (Citrus sinensis) plants but also facilitates Phytophthora and viral infections in Arabidopsis, and Nicotiana benthamiana (N. benthamiana). SDE3 directly associates with citrus cytosolic glyceraldehyde-3-phosphate dehydrogenases (CsGAPCs), which negatively regulates plant immunity. Overexpression of CsGAPCs and SDE3 significantly inhibits autophagy in citrus, Arabidopsis, and N. benthamiana. Intriguingly, SDE3 undermines autophagy-mediated immunity by the specific degradation of CsATG8 family proteins in a CsGAPC1-dependent manner. CsATG8 degradation is largely rescued by treatment with an inhibitor of the late autophagic pathway, E64d. Furthermore, ectopic expression of CsATG8s enhances Phytophthora resistance. Collectively, these results suggest that SDE3-CsGAPC interactions modulate CsATG8-mediated autophagy to enhance Las progression in citrus.
Abbreviations: ACP: asian citrus psyllid; ACD2: ACCELERATED CELL DEATH 2; ATG: autophagy related; Ca. Liberibacter: Candidatus Liberibacter; CaMV: cauliflower mosaic virus; CMV: cucumber mosaic virus; Cs: Citrus sinensis; EV: empty vector; GAPC: cytosolic glyceraldehyde-3-phosphate dehydrogenase; HLB: huanglongbing; H2O2: hydrogen peroxide; Las: liberibacter asiaticus; Laf: liberibacter africanus; Lam: liberibacter americanus; Pst: Pseudomonas syringae pv. tomato; PVX: potato virus X; ROS: reactive oxygen species; SDE3: sec-delivered effector 3; TEM: transmission electron microscopy; VIVE : virus-induced virulence effector; WT: wild-type; Xcc: Xanthomonas citri subsp. citri;
KEYWORDS: ATG8, E64D, effector, GAPC, plant immunity
Introduction
Citrus greening disease, also known as Huanglongbing (HLB), is a disastrous disease of citrus worldwide and is threatening the sustainability of the global citrus industry, especially in major citrus-growing regions of Asia and the Americas [1–3]. HLB is primarily caused by three “Candidatus Liberibacter” species, including “Ca. Liberibacter asiaticus” (Las), “Ca. Liberibacter africanus” (Laf), and “Ca. Liberibacter americanus” (Lam). The “Ca. Liberibacter” species are a group of fastidious phloem-limited bacteria transmitted by the feeding action of the Asian citrus psyllid (ACP), which blocks the flow of nutrients through vascular tissues, eventually leading to disease symptoms. Since its first report in China in the early last century, the incurable HLB disease has been documented in approximately 53 of 140 citrus fruit-producing countries in the world, accounting for the loss of billions of dollars annually in fruit yield and quality, tree damage, and management costs [1,4,5]. Current HLB control strategies include treatment with insecticides (to restrict ACP transmission) and the application of antibiotics or antimicrobial peptides (to limit Las growth) [6,7]; however, neither one of these strategies is effective in preventing HLB. Thus, the study of pathogenic mechanisms of “Ca. Liberibacter” species is urgently needed to cope with this devastating citrus disease and to ensure the survival of the global citrus industry.
Genome sequence analysis of Las, the most prevalent HLB pathogen worldwide, revealed its ability to secret effectors using a general Sec-dependent secretion apparatus [8]. Furthermore, 86 Sec-delivered effectors (SDEs) were confirmed to possess a functional Sec-secretion signal in Las. Among them, 21 are upregulated in planta and 5 are upregulated in psyllid host [9–11]. These SDEs are delivered to the phloem sieve cells and neighboring companion cells, and move through plasmodesmata [10,12,13]. This delivery mechanism is akin to the infection strategy used by Phytoplasmas, another class of insect-vectored phloem-limited bacterial pathogens, which also secrete effectors that move through plasmodesmata to modulate their host immunity [14]. Only two Las effectors have been identified to date: SDE1 (Las Sec-delivered effector 1) and SDE15. SDE1 shows high expression level in citrus and periwinkle hosts, inhibits plant defense by associating with citrus immune proteases, and causes leaf yellowing symptoms in transgenic citrus plants expressing SDE1 [12,15,16]. Las SDE15 interacts with ACD2 (ACCELERATED CELL DEATH 2), a red chlorophyll catabolite reductase, to suppress plant immunity and promote Las multiplication in citrus [17]. In Nicotiana benthamiana (N. benthamiana), several SDEs have been reported to suppress cell death induced by the pro-apoptotic mouse protein BAX and the Phytophthora infestans elicitin INF1 [18–20]. However, most Las SDEs have not been characterized, and it remains poorly understood how these SDEs promote Las colonization to contribute to the development of HLB symptoms in citrus.
GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is a classical metabolic enzyme that converts glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate in the glycolytic pathway, thereby generating energy and providing intermediates for cellular metabolism [21]. Because GAPDH is a housekeeping protein, its sequence is evolutionarily conserved across many organisms; however, only one isoform has been identified in animals, while plants possess multiple GAPDH isoforms [22]. In animals, GAPDH has been implicated in many cellular processes, including DNA repair, membrane fusion and transport, endocytosis and nuclear membrane assembly, and immune response, which are manipulated mainly by its oligomerization, post-translational modifications, and subcellular localization [23]. Similarly, plant GAPDHs interact with various proteins, exhibit diverse subcellular localization patterns, and play key roles in diverse cellular processes including macroautophagy/autophagy, mRNA regulation, redox sensing, immune response, and photosynthesis [24–29]. Several GAPDH genes have been identified in plant species including Arabidopsis, Nicotiana tobacco, common wheat (Triticum aestivum), rice (Oryza sativa), watermelon (Citrullus lanatus), and citrus (Citrus sinensis) [30]. The Arabidopsis genome encodes seven phosphorylating GAPDHs, including chloroplastic photosynthetic GAPDHs (GAPA1, GAPA2, and GAPB), cytosolic glycolytic GAPDHs (GAPC1 and GAPC2), plastidic glycolytic GAPDHs (GAPCp1 and GAPCp2), and one non-phosphorylating GAPDH, i.e., NP-GAPDH (NADP-dependent cytosolic GAPDH) [26,31]. The phosphorylating GAPDHs, sharing a highly reactive catalytic cysteine residue, can regulate multiple redox-induced post-translational modifications, and exhibit moonlighting properties in response to reactive oxygen species (ROS) [32]. Recent studies show that treatment with cadmium inactivates the GAPC enzyme and alters its subcellular localization from cytoplasm to nucleus, leading to cytoplasmic oxidation signaling in Arabidopsis [33]. Overexpression of GAPA1 in yeast and Arabidopsis suppressed ROS burst and triggered programmed cell death by the apoptosis regulator BAX [34]. Arabidopsis GAPCs bind to the plasma membrane-associated phospholipase D to transduce hydrogen peroxide (H2O2) signals in response to stress [35]. Additionally, GAPCs participate in diverse processes and impact plant innate immunity. Arabidopsis GAPC1 acts as a negative immune regulator against the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 [26]. In N. benthamiana plants challenged with Tobacco mosaic virus, Pst, or P. syringae pv. tabaci, GAPCs negatively regulate autophagy-mediated immunity by directly interacting with ATG3 (autophagy-related 3) [25]. Moreover, the Cotton leaf curl Multan virus βC1 protein binds to GAPCs and disrupts their interaction with ATG3, thus activating autophagy-induced immunity [36]. Although a total of six GAPDH genes have been identified in sweet orange [30], their roles in plant defense have not been fully explored.
Here, we identified a potential virulence factor SDE3 (CLIBASIA_00420) in Las-secreted proteins using a virus-induced virulence effector (VIVE) screening assay [37]. We show that SDE3 is conserved across six species in the Rhizobiaceae family. Overexpression of SDE3 effector facilitates Ca. Liberibacter asiaticus and Xanthomonas citri subsp. citri (Xcc) progression in citrus, Phytophthora infection in N. benthamiana, and Pst colonization in Arabidopsis. We demonstrate that SDE3 directly binds to the cytosolic CsGAPC1 and CsGAPC2 proteins, which function as a susceptibility factors to specifically suppress CsATG8-mediated immunity.
Results
Identification of SDE3 as a potential virulence effector in Las
Since “Ca. Liberibacter” species are not amenable to genetic manipulation and physiological studies, we employed a rapid functional screening assay to identify potential virulence proteins in Las [37]. We analyzed 23 putative secreted proteins (Table S1) that identified by Prasad et al. [10], and noticed that plants infiltrated with the PVX-CLIBASIA_00420 construct were dwarf and exhibited wrinkling and necrosis on the newly emerged upper leaves, whereas those inoculated with the PVX construct (control) exhibited mild viral symptoms (Figure 1A). Consistently, northern blot analysis showed that PVX genomic and subgenomic RNAs were accumulated to much higher levels in plants infiltrated with PVX-CLIBASIA_00420 than in those infiltrated with PVX (Figure 1B). CLIBASIA_00420 expression at the transcriptional level was confirmed in infiltrated N. benthamiana plants by reverse transcription-PCR (RT-PCR) (Fig. S1A). Those data indicate that CLIBASIA_00420 encodes a potential virulence effector and functions in promoting PVX virus accumulation and enhancing PVX infection symptoms. Moreover, RT-PCR analysis showed that CLIBASIA_00420 was highly expressed in an infected citrus tree (Fig. S1B). We therefore designated CLIBASIA_00420 as SDE3 (Sec-delivered effector 3), based on previous studies [16,17]. To further explore the virulence function of SDE3, we transiently expressed the SDE3 gene in N. benthamiana leaves. Leaves expressing SDE3 showed enhanced susceptibility to Phytophthora capsici (P. capsici) in the pathogen inoculation assay (Figure 1C), and P. capsici lesion length and relative biomass were markedly greater on leaves expressing SDE3 than on those transformed with the empty vector (EV) control (Figure 1D). These results suggest that SDE3 is a virulence factor that is essential for facilitating infection by virus and Phytophthora pathogens.
Figure 1.

Secretory protein of the HLB-associated pathogen, SDE3, is required for infection in N. benthamiana plants. (A) Disease symptoms of Nicotiana benthamiana (N. benthamiana) plants (n = 10) agroinfiltrated with the PVX construct (control) or recombinant PVX with 35S promoter-driven SDE3. Uninfected plants (mock) were used as negative controls. Pictures were taken 21 d after agroinfiltration. (B) Northern blots showing the accumulation of PVX genomic and subgenomic RNAs at 21 days post-inoculation (dpi). (C) Symptoms of Phytophthora capsici (P. capsici) infection on N. benthamiana leaves (n = 15) expressing the empty vector (EV; control) or SDE3. Leaves were photographed under UV light at 48 hours post-inoculation (hpi). Scale bars: 0.9 cm. (D) Analysis of lesion size and relative biomass on N. benthamiana leaves caused by P. capsici. Relative biomass was determined by genomic DNA (gDNA)-based quantitative PCR (qPCR). Data represent mean ± standard error (SE). Asterisks indicate statistically significant differences (P < 0.01; Duncan’s multiple range test). (E) Phylogenetic analysis of SDE3-like proteins in bacteria. Neighbor-joining tree of 19 SDE3-like proteins was constructed using full-length amino acid sequences. The clade containing SDE3-like proteins of Candidatus Liberibacter is highlighted in green, and SDE3 is highlighted in bold red font. All experiments were performed in triplicate, with similar results.
Apart from its SP, SDE3 does not display any significant feature; it encodes an approximately 16 kDa protein, and mainly localizes to the nucleus and cytoplasm in the epidermal cells of N. benthamiana leaves (Figure S1C-E). Interestingly, the amino acid sequence of Las SDE3 homologs is highly conserved among the five “Ca Liberibacter” species, with 51.5–64.8% sequence identity, and 14 potential homologs of Las SDE3 were identified in bacteria belonging to six different genera of the Rhizobiaceae family (Figure 1E, S1F), suggesting that SDE3 is an evolutionarily conserved effector.
SDE3 expression promotes bacterial and Phytophthora infection in citrus and Arabidopsis
To further elucidate the biological role of SDE3 in plant immunity, we generated transgenic Arabidopsis lines expressing SDE3, the expression of SDE3 in the transgenic lines was confirmed by western blotting (Fig. S2A). Two independent transgenic lines (SDE3–16 and SDE3–18) showed no morphological changes or growth and developmental abnormalities relative to wild-type plants (Fig. S2B). We then subjected these transgenic lines to bacterial and Phytophthora infection assays. The growth of Pst DC3000 was significantly higher in the two transgenic lines than in wild-type plants (Figure 2A). Similar results were obtained when both transgenic lines infected with P. capsici were examined at 48 hours post-inoculation (hpi) (Figure 2(B–D)). Taken together, these results reveal an inhibitory role of SDE3 in modulating plant immunity in Arabidopsis.
Figure 2.

Overexpression of SDE3 promotes bacterial and Phytophthora infection in transgenic Arabidopsis and citrus plants. (A) Bacterial growth in transgenic Arabidopsis lines expressing the bacterial effector gene SDE3. Bacterial titers were evaluated at 0 and 3 dpi. (B) Evaluation of disease symptoms on transgenic Arabidopsis leaves by trypan blue staining. Leaves of SDE3-overexpressing Arabidopsis plants were inoculated with zoospores of P. capsici strain Pc35, and symptoms were evaluated at 48 hpi. Arabidopsis ecotype Columbia (Col; wild type) was used as a negative control. Scale bars: 0.6 cm. (C) Quantitative analysis of disease severity. **P < 0.01 (as determined by the Wilcoxon rank-sum test). (D) Relative biomass of P. capsici in Arabidopsis leaves, as analyzed by gDNA-based qPCR. (E) Citrus canker symptoms on the leaves of SDE3-overexpressing plants and EV plants inoculated with Xanthomonas citri subsp. citri (Xcc). Assays were performed in vitro using the pinprick inoculation method. One microliter of the Xcc suspension (1 × 108 CFU/ml) was loaded onto each pinprick. Uninfected citrus leaves were used as controls. (F) Bacterial growth in leaves of transgenic SDE3 and WT plants at 9 dpi. ND, Not detected. (G) “Ca. L. asiaticus” bacterial titer in the SDE3-transgenic citrus and EV-transgenic control citrus were determined by quantitative PCR during a 1-year period after graft inoculation. The Ct value of each amplicon represents the “Ca. L. asiaticus” genomic copy numbers in 100 ng of DNA. Data in (A, D, F, and G) represent mean ± SE. Different letters indicate statistically significant differences (P < 0.01; Duncan’s multiple range test). The experiment was performed twice, with similar results.
The ability of SDE3 to subvert plant immunity prompted us to determine whether its expression can facilitate the bacterial pathogen infection in citrus plants. We overexpressed the SDE3 gene in sweet orange cultivar “Wanjincheng” via Agrobacterium tumefaciens-mediated epicotyl transformation, and generated two stable transgenic citrus plants (SDE3–1 and SDE3–2) that was confirmed by RT-PCR analysis (Fig. S2C). Similar to that in transgenic Arabidopsis lines, we did not find the developmental abnormalities in SDE3-overexpressing citrus plants (Fig. S2D). Citrus canker, caused by Xcc, is serious bacterial disease that causes lesions on the leaves, stems, and fruit of plants [38]. We firstly challenged two lines (SDE3–1 and SDE3–2) with Xcc using the pinprick method. We noticed that canker-like bulges appeared earlier on SDE3-overexpressing leaves than that on EV (control) leaves after Xcc inoculation, indicating a faster growth rate of Xcc in SDE3-overexpressing lines compared with the control (Figure 2E). Additionally, at 9 dpi, the Xcc titer was significantly higher in SDE3-overexpressing leaves than in EV leaves (Figure 2F).
To determine the susceptibility of SDE3 transgenic citrus to “Ca. L. asiaticus”, two independent SDE3-expressing citrus lines (1 year old) were graft inoculated using buds from “Ca. L. asiaticus”-infected Valencia sweet orange plants. As a control, wild-type sweet orange plants haboring EV at the same age were also graft inoculated. Quantitative reverse transcription PCR (qRT-PCR) was used to monitor “Ca. L. asiaticus” titers (Figure 2G). We successed to graft Las onto SDE3–1 plant, but failed to graft Las onto SDE3–2 plant. Intriguingly, comparison of the cycle threshold (Ct) values found that SDE3–1 plant was more susceptible to “Ca. L. asiaticus” than the control plants (Figure 2G). These results strongly suggest that SDE3 promotes “Ca. L. asiaticus” colonization and proliferation in citrus. Overall, these results indicate that SDE3 acts as a virulence factor that promotes citrus susceptibility to both HLB and canker, two severe citrus diseases worldwide.
SDE3 physically associates with CsGAPC1 and CsGAPC2
To reveal the virulence function of SDE3, the yellow fluorescent protein (YFP) gene- and FLAG-tagged SDE3 gene (YFP-SDE3-3*FLAG) was transiently expressed in N. benthamiana leaves, and proteins were immunoprecipitated using anti-FLAG affinity beads. The recovered proteins were then analyzed by liquid chromatography – tandem mass spectrometry (LC – MS/MS) to identify the interacting partners of SDE3. Totally 18 proteins were identified with FDR ≤0.01 as threshold (Table S2). Among them, N. benthamiana cytosolic glycolytic GAPDHs NbGAPC1 was selected for further analysis, based on its roles in plant autophagy-mediated immunity [25] and two NbGAPCs were identified in LC-MS analysis. Sequence analysis revealed that NbGAPCs has two homologs in the citrus genome (CsGAPC1/Cs5g06870 and CsGAPC2/Cs7g10220), and the NbGAPC1 protein shares 90.2–95.9% amino acid sequence identifies with CsGAPC1 and CsGAPC2 (Fig. S3). Since GAPCs belong to the GAPDH gene family, a phylogenetic tree of 27 GAPDH genes from Arabidopsis, N. benthamiana, Rice, and Citrus was construced (Fig. S4A), CsGAPC1 and CsGAPC2 were unsurprisingly clusted into a group with GAPC proteins from other three species, indicating that GAPCs are relatively conserved proteins.
To validate the association of SDE3 with CsGAPC1 and CsGAPC2, we conducted the bimolecular fluorescence complementation (BiFC) assay to confirm the physical interaction of SDE3 with CsGAPC1 and CsGAPC2. The SDE3, CsGAPC1 and CsGAPC2 genes were fused to the YFP gene sequence corresponding to its N-terminal half (YN) and C-terminal half (YC), respectively, and the resulting constructs were coexpressed in N. benthamiana leaves. At 48 hpi, strong YFP signal was detected in the epidermal cells of leaves expressing CsGAPC1-YC and CsGAPC2-YC together with SDE3-YN but not in cells expressing CsGAPC1-YC+YN, CsGAPC2-YC+YN or YC+SDE3-YN (Figure 3A). The YFP signal was mainly localized to the cytoplasm. We then verified the interaction between SDE3 and CsGAPC1, SDE3 and CsGAPC2 by performing an in vitro affinity-isolation assay, respectively. When the purified SDE3 fused to six histidine (His) residues was incubated with maltose-binding protein (MBP)-tagged CsGAPC1 and CsGAPC2, the SDE3-His protein was specifically enriched in MBP-bound resins (Figure 3B). In addition, we also performed a co-immunoprecipitation (co-IP) assay. The YFP-SDE3 or YFP-3*HA fusion was transiently coexpressed with CsGAPC1–3*FLAG, CsGAPC2–3*FLAG in N. benthamiana leaves via agroinfiltration. Total proteins were extracted from the infiltrated leaves and incubated with anti-GFP resin. Immunoblotting with anti-FLAG antibody revealed remarkable enrichment of YFP-SDE3, but not that of YFP-3*HA, in the precipitate (Figure 3C). Collectively, these results demonstrate the association of SDE3 with CsGAPC1 and CsGAPC2 both in vitro and in vivo.
Figure 3.

SDE3 targets CsGAPC1 and CsGAPC2 proteins both in vitro and in vivo. (A) Bimolecular fluorescence complementation (BiFC) assays in the epidermal cells of N. benthamiana leaves. The cYFP and nYFP empty vectors were served as negative controls in each combination. Fluorescence indicating SDE3-CsGAPC1, SDE3-CsGAPC2 and SDE3-CsGAPC1Cter interactions was detected by confocal microscopy at 48 hpi. Scale bars: 30 μm. (B) in vitro affinity-isolation assay. His-SDE3, MBP-CsGAPC1, MBP-CsGAPC2, MBP- CsGPAC1Nter, and MBP-CsGAPC1Cter were expressed in Escherichia coli. MBP-tagged CsGAPC1 and CsGAPC2 or their truncated variants-bound MBP beads were incubated with the purified His-SDE3 protein. Co-precipitation of SDE3 with CsGAPC1 and CsGAPC2 and CsGAPC1Cter was examined by western blotting using specific antibodies. (C) Co-immunoprecipitation (co-IP) assay confirming that SDE3 co-precipitates with citrus CsGAPC1 and CsGAPC2 proteins, respectively. Total proteins were extracted from N. benthamiana leaves expressing the YFP-SDE3, YFP-3*HA, CsGAPC1–3*FLAG, or CsGAPC2–3*FLAG construct. The immune complexes were immobilized on anti-GFP magnetic beads, and the co-precipitation of CsGAPC1 and CsGAPC2 was examined by western blotting using specific antibodies. (D) Schematic representation of the two CsGAPC1 truncations. The conserved NAD-binding domain and catalytic domain are indicated. (E) SDE3 induces accumulation of CsGAPC1 proteins in N. benthamiana. CsGAPC1 was transiently coexpressed with SDE3 or YFP in N. benthamiana leaves, respectively. Immunodetection of expression of proteins in using anti-FLAG, anti-GFP, and anti-Actin antibody, Actin expression as a loading control for western blot analysis. The experiments were performed twice, with similar results.
Amino acid sequence analysis of the CsGAPC proteins revealed the presence of a NAD-binding domain at its N terminus (CsGAPC1Nter) and a catalytic domain at its C terminus (CsGAPC1Cter) (Figure 3D). In order to verify the binding domain of CsGAPCs with SDE3, two CsGAPC1 mutants, CsGAPC1Nter and CsGAPC1Cter, was constructed. Intriguingly, in vitro affinity-isolation assay showed that CsGAPC1Nter, but not CsGAPC1Cter can be detected in His precipitate (Figure 3B). This result was further verified in the epidermal cells of N. benthamiana leaves by the BiFC assay (Figure 3A), suggesting that the NAD-binding domain of CsGAPC1 is required for their interaction.
Given that SDE3 associates with CsGAPCs, we examined the effect of SDE3 on CsGAPC protein. CsGAPC1 was transiently co-expressed with SDE3 or EV in N. benthamiana. Western blot results revealed that the enhanced accumulation of CsGAPCs protein was detected in leaves coexpressed with SDE3 compared wtih those coexpressed with EV leaves (Figure 3E), indicating that SDE3 may likely induce CsGAPCs protein accumulation.
CsGapcs negatively regulate plant immunity
Next, we investigated the tissue-specific expression pattern of CsGAPC1 and CsGAPC2 in citrus by qRT-PCR. The results showed that CsGAPC1 and CsGAPC2 were expressed ubiquitously in all citrus organs tested, with the highest expression in leaf veins, and relatively low expression in roots and leaf mesophyll tissues (Fig. S4B). Subsequently, we determined the subcellular localization of CsGAPCs by transiently expressing the CsGAPC1/-GFP or CsGAPC2-GFP constructs in N. benthamiana leaves via Agrobacterium-mediated transformation. Confocal microscopy analysis of the agroinfiltrated leaves showed that both proteins localized primarily to the cytoplasm of leaf epidermal cells (Fig. S4C, S4D). Similarly, two truncated derivates of CsGAPC2 (CsGAPC2Nter and CsGAPC2Cter) also showed cytosolic localization (Fig. S4C), indicating that truncation of the N- or C-terminal half of the CsGAPC2 protein does not alter its subcellular localization.
To elucidate the potential role(s) of CsGAPCs in plant immunity, transgenic Arabidopsis lines (CsGAPC1–4 and CsGAPC1–29) expressing CsGAPC1 were generated. Neither of the transgenic lines exhibited any visible morphological defects, and their growth and development were reminiscent of those of wild-type plants (Fig. S2A, S2B). Both CsGAPC1-overexpressing lines were more susceptible to the bacterial pathogen Pst, and showed greater bacterial growth than wild-type plants (Figure 4A). Similar results were obtained when these two transgenic lines were examined at 48 hpi upon P. capsici infection (Figure 4B, C). Consistent with the above data, transient expression of CsGAPC1 or CsGAPC2 in N. benthamiana leaves enhanced susceptibility to P. capsici as indicated by the relative biomass and lesion length of the Phytophthora pathogen, which were significantly greater on leaves transiently expressing CsGAPC1 or CsGAPC2 than on those transformed with the GFP-containing EV control.
Figure 4.

CsGAPC expression negatively regulates plant defense in Arabidopsis and N. benthamiana. (A) Growth of Pst on the leaves of transgenic Arabidopsis lines expressing the CsGAPC1 gene. Bacterial titers were evaluated at 0 and 3 dpi. (B) Symptoms of P. capsici infection on leaves (n = 15) expressing the CsGAPC1 gene. Leaves were photographed under white light at 48 hpi. Scale bars: 0.4 cm. (C) Lesion size and relative biomass of P. capsici. Symptoms of P. capsici infection (D) and its lesion size and relative biomass (E) on NbGAPCs-silenced N. benthamiana leaves. Symptoms of P. capsici infection (F) and its lesion size and relative biomass (G) on NbGAPCs-silenced leaves expressing SDE3. In (C, E and G) relative biomass of the pathogen was determined by gDNA-based qPCR. For relative biomass quantification, the ratio of P. capsici DNA compared with Arabidopsis (C) or N. benthamiana (E and G) DNA was measured. Scale bars in (D and F): 0.9 cm. Data in (A, C, E, and G) represent mean ± SE. Different letters indicate statistically significant differences (P < 0.01; Duncan’s multiple range test). The experiment was performed twice, with similar results.
To further explore the crucial domain for their roles in plant immunity, CsGAPC2Nter and CsGAPC2Cter were transiently expressed in N. benthamiana leaves, and then subjected to P. capsici infection. Results showed that the lesion size and relative biomass of leaves expressing CsGAPC2Nter were much lower than that of CsGAPC2Cter and similar to that of EV (Fig. S5A-C), indicating that the NAD-binding domain is also in charge of CsGAPCs roles in plant immunity.
Next, we investigated GAPC homologs in N. benthamiana using virus-induced gene silencing (VIGS) to determine their function in plant defense during Phytophthora infection. Two VIGS constructs (VIGS1 and VIGS2) were generated to concurrently silence three NbGAPC genes. Each VIGS construct successfully silenced these three targets as shown by qRT-PCR analysis, without affecting the growth and development of N. benthamiana plants (Fig. S5D, E), and the NbGAPC-silenced plants were more resistant to P. capsici than the control plants (Figure 4D, E). Importantly, we found that expression of SDE3 was not able to promote Phytophthora infection in NbGAPC-silenced leaves compared with GFP-silenced leaves, indicating that SDE3 depends on NbGAPCs to inhibit the innate immunity of N. benthamiana plants (Figure 4F, G; S5F). Collectively, these data suggest that CsGAPC1 and CsGAPC2 act as negative immune regulators in citrus. This result is in agreement with previous findings showing that both NbGAPCs function as susceptibility factors to facilitate infection [25,26].
SDE3 inhibits CsGAPC1-dependent autophagy in plants
Since GAPC genes have been reported to negatively regulate autophagy-mediated immunity in N. benthamiana and Arabidopsis [25,26], we examined whether CsGAPCs also participate in manipulating autophagy in citrus. Primary roots of 7–8-day-old CsGAPC1-overexpressing and wild-type (Col) Arabidopsis plants were treated with benzo-(1, 2, 3)-thiadiaole-7-carbothioic acid (BTH; autophagy activator), and then stained with monodansylcadaverine (MDC; a fluorescent dye that specifically binds to autophagic bodies). Compared with Col roots, the roots of both transgenic lines (CsGAPC1–4 and CsGAPC1–29) exhibited reduced accumulation of MDC-labeled autophagic bodies, with 2–3-fold times lower levels of autophagy activity (Figure 5A, B). Plant NBR1, a selective autophagy cargo receptor, could be accumulated in the vacuole when autophagy activity is reduced [39,40]. Therefore, the expression level of NBR1 was examined in Col and two CsGAPC1 transgenic lines. Western blot results showed that more NBR1 protein was accumulated in CsGAPC1–4 and CsGAPC1–29 transgenic lines than that in Col, while lower NBR1 expression was detected in atgapc1 and atgapc2 mutants (Figure 5C). Taken together, those results above indicated that CsGAPC1, like NbGAPC1 and AtGAPC1, also suppresses autophagy activity in transgenic Arabidopsis plants.
Figure 5.

Autophagy is suppressed in SDE3- and CsGAPC1-overexpressing Arabidopsis and N. benthamiana plants and the suppression by SDE3 is dependent on AtGAPC in plants. (A) Representative confocal microscopy images of autophagic activity in Col, CsGAPC1 and SDE3 overexpressing Arabidopsis plants. Autophagic bodies were revealed by monodansylcadaverine (MDC) staining. Scale bars: 20 μm. Lower panels showing enlarged areas enclosed by red squares in top panels. (B) Relative autophagic activity in CsGAPC1 and SDE3-overexpressing transgenic Arabidopsis roots. (C) Western blotting analysis showing NBR1 accumulation in SDE3, CsGAPC1 transgenic, atgapc1 and atgapc2 mutant Arabidopsis lines, Rubisco was used as loading controls. (D) Representative transmission electron microscopy (TEM) images of autophagic structures. The ultrastructure of autophagic bodies was observed in the vacuoles (V) of mesophyll cells of Col (control) and SDE3-overexpressing plants. Red arrows represent the autophagic bodies. (E) Relative autophagic activity in SDE3-overexpressing Arabidopsis roots and NbATG8F coexpressed with CsGAPC1 or CsGAPC1 and SDE3 N. benthamiana leaves. (F) Representative TEM images of autophagic structures. The ultrastructure of autophagic bodies was observed in the vacuoles (V) of mesophyll cells of “Wanjincheng” orange tree (EV) and SDE3-overexpressing orange tree SDE3–2. (G) Relative autophagic activity in BTH-activated SDE3-overexpressing transgenic citrus leaves. (H) Representative confocal microscopy images of autophagic activity in Col, atgapc1, atgapc2 mutants and atgapc1/SDE3–4, atgapc1/SDE3–10 plants. Autophagy was revealed by MDC staining. (I) Relative autophagic activity in Col, atgapc1, atgapc2 mutant and atgapc1/SDE3–4, atgapc1/SDE3–10 plants. In (A, D, F and H), all the samples subjected for confocal microscopy and TEM analysis were treated with BTH for autophagy activation. In (B, E, G and I) values obtained in transgenic or mutant plants were normalized to that obtained in Col or Wanjincheng plants, which was set to 1.0. The autophagic puncta were quantified in approximately 20–30 cells in each treatment. Data represent mean ± SE. Different letters indicate statistically significant differences (P < 0.01; Duncan’s multiple range test). The experiment was performed twice, with similar results. In (D and F), Red arrows represent the autophagic bodies. CW, cell wall; Cp, chloroplast; M, mitochondria.
Given that SDE3 associates with CsGAPCs, we investigated whether SDE3 plays a direct role in autophagy. We monitored autophagy activity in SDE3-expressing transgenic Arabidopsis plants using the MDC staining approach. Approximately 3–4-fold times fewer autophagic bodies were observed in SDE3 transgenic plants than in non-transgenic Col plants (Figure 5A, B). A similar level of reduction was observed when we further investigated the effect of SDE3 on autophagy in the vacuole of SDE3 transgenic Arabidopsis plants using transmission electron microscopy (TEM) (Figure 5D, E). Consistent with that in SDE3-transgenic Arabidopsis, the number of autophagic bodies were markdedly less in SDE3-2 transgenic citrus leaves (Since SDE-1 transgenic citrus tree has been infected with Las, only SDE-2 plant was used for TEM analysis for comparison with “Wanjincheng” orange) than that in citrus leaves harboring EV (Figure 5F, G). The accumulation of NBR1 protein was significantly increased in SDE3–16 and SDE3–18 trangenic plants (Figure 5C). Moreover, our data showed that the reduction of autophagic activity was also obtained in HLB-infected citrus plants compared with healthy citrus plants by TEM analysis (Fig. S5G, H). These results suggest that SDE3 expression suppresses autophagy in plants.
As both the SDE3 and CsGAPCs proteins showed the suppression of autophagy, this further prompted us to test whether the inhibition of autophagy by SDE3 expression is dependent on the function of GAPCs. Two SDE3 overexpressing transgenic Arabidopsis lines in atgapc1 mutant background, atgapc1/SDE3–4 and atgapc1/SDE3–10, were obtained and the SDE3 expression was verified by western blot (Fig. S5I). MDC-labeled autophagic bodies were examined in BTH treated primary roots of Col, atgapc1, atgapc2, atgapc1/SDE3–4 and atgapc1/SDE3–10, and no reduction was observed in atgapc1, atgapc2 mutants and SDE3 expression did not result in the reduced accumulation of autophagic bodies in atgapc1/SDE3–4 and atgapc1/SDE3–10 plants. (Figure 5H, I). These results suggest that the autophagy inhibition by SDE3 is dependent on GAPC expression.
SDE3 aggravates the suppression of CsGAPC1-mediated autophagy through the specific degradation of CsATG8 family proteins
To further confirm the effect of the SDE3-CsGAPC1 interaction on autophagy, we firstly investigated the subcellular localization of NbATG8F when the NbATG8F gene was coexpressed with EV, SDE3 and/or CsGAPC1. NbATG8F is a well-characterized autophagy-related protein that localizes to the nucleus and cytoplasm [41]. The CsGAPC1 protein relocalized part of NbATG8F from the nucleus to the cytoplasm, and weak nuclear signal was detected in the epidermal cells of N. benthamiana leaves, while SDE3 did not alter the subcellular localization of NbATG8F (Figure 6A). Interestingly, this altered localization of NbATG8F by CsGAPC1 was further exacerbated when NbATG8F was coexpressed with both CsGAPC1 and SDE3; only weak fluorescent signal of NbATG8F was detected (Figure 6A). We further confirm the effect of both SDE3 and CsGAPC1 on NbATG8F by the fraction of the nuclear and cytoplasmic protein extracts together with western blot analysis (Figure 6B).
Figure 6.
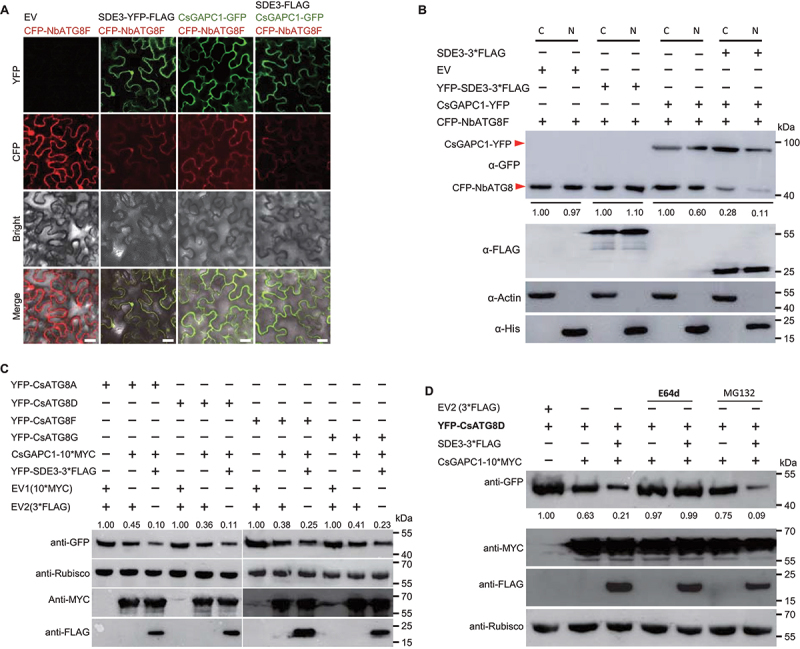
SDE3 suppresses CsGAPC1-mediated autophagy by altering the stability and localization of CsATG8s. (A) Subcellular localization analysis of SDE3, NbATG8F, and CsGAPC1 proteins in the epidermal cells of N. benthamiana leaves. All images were captured using a confocal fluorescence microscope at 48 hpi. Scale bars: 30 μm. (B) Immunoblot analysis showing that SDE3-CsGAPC1 interaction impedes the subcellular localization of NbATG8F. Constructs were transiently coexpressed in N. benthamiana leaves by agroinfiltration and CFP-NbATG8F accumulation in nuclear and cytoplasmic fractions were revealed by western blot, respectively, in 48 hpi. C, cytoplasmic fraction of transiently expression plant cells. N, nuclear fraction of transiently expression plant cells. Histone and Rubisco were used as the nuclear and cytoplasmic markers, respectively. (C) Immunoblot analysis showing that SDE3 exacerbates the expression inhibition of four autophagy-related CsATG8 paralogs by CsGAPC1. Constructs were transiently expressed or coexpressed in N. benthamiana leaves. Protein accumulation was examined by western blot analysis using specific antibodies. (D) Inhibition of the late autophagic pathway with E64d largely inhibits the degradation of CsATG8D by CsGAPC1. CsATG8D together with CsGAPC1 and/or SDE3 were transiently coexpressed in N. benthamiana leaves via agroinfiltration, and the infiltrated leaves were treated with E64d, or MG132 at 24 hpi and examined at 48 dpi. Protein accumulation was examined by western blot analysis using specific antibodies. Rubisco was used as a loading control in western blot analysis. The experiment was performed twice, with similar results.
The results of subcellular localization seen when co-expressing NbATG8F with CsGAPC1 or SDE3 and CsGAPC1 led us to further verify if SDE3 and CsGAPCs have a direct effect on autophagy. We cloned and transiently expressed CsATG8 alleles in N. benthamiana leaves. Western blotting results showed that CsGAPC1 induced the degradation of CsATG8A, and the effect of CsGAPC1 on CsATG8A degradation increased considerably in the presence of SDE3; nonetheless, expression of CsGAPC1 and SDE3 did not lead to the total disappearance of the CsATG8A protein (Figure 6C). Similar results were obtained when CsGAPC1 was coexpressed with three other CsATG8 alleles (Figure 6C). We then used quantitative real-time PCR to analyze three CsATG8s transcript levels, and found that CsATG8s expression was not significantly altered when CsATG8s were coexpressed with CsGAPC1 and EV or CsGAPC1 and SDE3 (Fig. S6A). We also determined two ATG8s transcript levels (AtATG8C and AtATG8I) in SDE3 or CsGAPC1 transgenic Arabidopsis plants, and also found that SDE3 and CsGAPCs expression have no obvious effect on AtATG8s transcript levels (Fig. S6B). Therefore, it is likely that the SDE3-CsGAPC interaction induced reduction in CsATG8s occurs at the protein level. Collectively, these data further support our hypothesis that SDE3 modulates the CsGAPC1-mediated protein stability of CsATG8s, thereby ihibiting autophagy in plants.
To explore the possible mechanism of CsATG8 degradation, CsATG8D or CsATG8G was transiently coexpressed either with CsGAPC1 and EV or with CsGAPC1 and SDE3 in N. benthamiana leaves, and the infiltrated leaves were treated with a couple of protease inhibitors. The application of MG132, a protease inhibitor known to block the proteolytic activity of the 26S proteasome complex, did not prevent the degradation of CsATG8D and CsATG8G (Figure 6D; S6C). To further elucidate the role of autophagy in the SDE3–CsGAPC1 interaction, we used E64d, an inhibitor of the autophagic degradation pathway (Figure 6D; S6C). By contrast, application of E64d largely stabilized both CsATG8D and CsATG8G proteins, despite the presence of SDE3 (Figure 6D; S6C). Taken together, our results indicate that the SDE3-CsGAPC1 interaction induces the degradation of CsATG8s, probably via the autophagy pathway.
These data prompted us to explore the functions of CsATG8s. We firstly investigated the effect of CsATG8s on autophagy by detecting expression of NBR1 protein. The results showed that expression of three CsATGs (CsATG8D, CsATG8F and CsATG8G) remarkably inhibited the accumulation of NBR1 protein (Fig. S6D). We then performed a cleavage assay of free GFP in N. benthamiana and validated whether CsATG8F is involved in the autophagy process [42–44]. YFP-CsATG8F–3*FLAG was transiently expressed in N. benthamiana. We found that YFP tag could be cleaved from YFP-CsATG8F–3*FLAG fusion protein upon the BTH treatment. And CsATG8F was markedly degraded in the presence of CsGAPC1 alone or CsGAPC1 and SDE3, ultimately only free GFP was detected (Fig. S6E), indicating that CsATG8F is a functional autophagic protein and the SDE3 and CsGAPC interaction-mediated degradation might happen during the autophagic pathway. Finally, we evaluated the function of CsATG8s in plant defense, P. capsici infection assay was performed. Intriguingly, the ectopic expression of three CsATG8 alleles markedly suppressed infection against P. capsici in N. benthamiana (Fig. S6F – S6H), suggesting that CsATG8s may act as a positive regulator of immunity in plants.
CsAtg8s and CsATG3 are directly targeted by CsGAPC1
Having established that the SDE3-CsGAPC1 interaction regulates CsATG8-mediated autophagy in citrus, we determined whether CsGAPC1 directly associates with CsATG8 proteins. We cloned six CsATG8 alleles, and conducted the BiFC assay by coexpressing CsGAPC1-YC and CsATG8s-YN constructs in N. benthamiana leaves. At 48 hpi, strong YFP signals were detected in leaf epidermal cells expressing CsGAPC1-YC+CsATG8s-YN but not in cells expressing CsGAPC1-YC+YN or YC+CsATG8s-YN (Fig. S7A). The fluorescence signals were mainly detected in the nucleus and cytoplasm. Next, we chose three CsATG8 proteins (CsATG8F, CsATG8G, and CsATG8I) and carried out in vitro affinity-isolation assays in E. coli expressing CsATG8s-His and MBP-CsGAPC1. Our data showed that the three CsATG8s-His proteins were specifically enriched in MBP-CsGAPC1-bound amylose resins (Figure 7A). We further confirmed the CsGAPC1-CsATG8s interactions in planta using co-IP assays. CsGAPC1–3*FLAG was transiently coexpressed with each YFP-CsATG8F construct or with YFP-3*HA in N. benthamiana leaves via agroinfiltration. Total proteins were extracted from the infiltrated leaves and incubated with anti-GFP resin. The CsGAPC1–3*FLAG proteins, but not YFP-3*HA, were remarkably enriched in the YFP-CsATG8F precipitate. Similarly, CsGAPC1–3*FLAG, but not YFP-3*HA, were markedly enriched in the YFP-CsATG8G and YFP-CsATG8I precipitates with anti-GFP resin (Figure 7B). Collectively, these results demonstrated the direct interaction of CsGAPC1 with CsATG8s.
Figure 7.

CsGAPC1 specifically associates with the autophagic proteins, CsATG8s and CsATG3. (A) In vitro affinity-isolation assay. MBP, MBP-CsGAPC1, and His-tagged CsATG8 paralogs were expressed in E. coli. MBP-CsGAPC1-bound beads were incubated with the purified His-tagged CsATG8 paralogs. Co-precipitation of MBP-CsGAPC1 with three CsATG8 proteins (CsATG8F/G/I) were examined by western blotting using specific antibodies. (B) Co-IP experiments confirming that CsGAPC1 co-precipitates with the three CsATG8 paralogs. Total proteins were extracted from N. benthamiana leaves expressing the CsGAPC1–3*FLAG, YFP-3*HA, or YFP-CsATG8F/G/I construct. The immune complexes were immobilized using anti-GFP magnetic beads, and co-precipitation of CsGAPC1 was examined by western blotting using specific antibodies. (C) Co-IP assay displaying that CsGAPC1 co-precipitates with CsATG3. Total proteins were extracted from N. benthamiana leaves expressing YFP-CsGAPC1, YFP-3*HA, or CsATG3–3*FLAG constructs and immobilized by anti-FLAG magnetic beads. Co-precipitation of CsGAPC1 was examined by western blotting using specific antibodies. (D) CsGAPC1 did not affect expression of two citrus autophagy-related proteins in N. benthamiana. CsGAPC1 was transiently coexpressed with CsTAG3, CsTAG6, CsATG1A or CsATG9 in N. benthamiana leaves, respectively. Immunodetection of three CsATGs proteins used anti-FLAG, anti-GFP, or anti-Actin antibody, Actin expression as a loading control for western blot analysis. The experiments in (A, B, C and D) was performed twice, with similar results. (E) Proposed model showing the role of SDE3 in regulating Las progression in citrus. During Ca. Las infection, the virulence effector SDE3 of “Ca. Liberibacter asiaticus” interacts with susceptibility factor CsGAPCs in citrus. By the direct interaction of CsGAPCs with CsATG8s, SDE3 eventually aggravates the specific degradation of CsATG8 family proteins and inhibits the formation of autophagic bodies, thereby inhibiting CsATG8-mediated immunity and resulting in the disease development of HLB in citrus.
Previous results showed that NbGAPC1 can directly associate with NbATG3 [25]. To verify this interaction in citrus, we performed a Co-IP assay. The results exhibited that CsATG3–3*FLAG was detected in CsATG3–3*FLAG precipitate, but not in YFP-3*HA precipitate (Figure 7C), suggesting that CsATG3 is also a target of CsGAPCs. These data led us to investigate whether the SDE3-CsGAPC1 interaction has a common or specific side effect on other autophagy-related proteins in citrus. We chose four genes (CsATG1A, CsATG3, CsATG6 and CsATG9) involved at different steps of the autophagic body formation. We next generated the expressing constructs of these genes (CsATG1A-3*FLAG, CsATG3–3*FLAG, CsATG6–3*FLAG and CsATG9-3*FLAG), each one of which was then transiently coexpressed in N. benthamiana leaves with CsGAPC1 or YFP. Western blotting showed that all CsATG proteins accumulated to similar levels in N. benthamiana leaves, regardless of the presence or absence of CsGAPC1 (Figure 7D), indicating that CsGAPC proteins specifically inhibit the stability of CsATG8 proteins.
In combination with the ATGs protein degradation and CsGAPCs targets, we can conclude that CsATG8s might not the only targets of CsGAPCs in plants, however CsGAPCs have the effect of specific degradation on CsATG8s, but not other CsATG proteins.
Discussion
HLB is the most catastrophic disease threatening citrus production globally. The complexity of Las-psyllid-Citrus interactions, owing to the inability to culture Las, unpredictable behavior and ecology of the insect vector, and complex and perennial nature of the host plant, has led to many technical challenges [5,45]. Given that a single technological breakthrough is unlikely to combat the citrus greening disease, it is crucial for researchers to continue broad efforts in multiple disciplines. In line with this idea, we recently developed a functional screening assay (VIVE) using PVX-based expression vector, which can be used to identify potential virulence factors in unculturable pathogens [37]. In the current study, we investigated the virulence mechanism underlying a sec-dependent secreted protein, and identified SDE3 as a potential virulence effector in Las. Subsequently, using SDE3 as a molecular probe, we identified CsGAPCs as its virulence partner, thereby offering mechanistic insights into the virulence strategies employed by SDE3 during “Ca. L. asiaticus” phloem colonization and HLB disease progression.
Cytoplasmic GAPCs have been shown to relocalize to the plant cell nucleus under oxidative stress, for example, upon exposure to H2O2 [29,46]. Similarly, strong nuclear accumulation of AtGAPCs was reported during giant cell formation, and the corresponding proteins were targeted by Meloidogyne incognita nuclear effector MiEFF1 [47]. However, the function of GAPCs in the nucleus remains unclear. In this study, we found that the subcellular localization of CsGAPC1 protein does not alter in the presence and absence of SDE3. Conversely, CsGAPC1 induces the localization alteration of NbATG8F and seems to affect its stability in N. benthanmiana leaves, and this effect was further aggregated when CsGAPC1 coexpressed with SDE3 in both cytoplasm and nucleus of N. benthanmiana leaves, indicating that CsGAPC1 and SDE3 execute their functions in the cytoplasm and nucleus. Consistently, SDE3 suppresses immune responses in citrus by interacting with CsGAPC1 and CsGAPC2, the homologs of Arabidopsis AtGAPC1 and AtGAPC2 and N. benthamiana NbGAPC1, NbGAPC2 and NbGAPC3 in the cytoplasm. Both AtGAPC and NbGAPC have been reported to negatively regulate plant immunity [25,26]. Intriguingly, similar results that CsGAPCs function in plant defense were obtained when CsGAPC genes were overexpressed in Arabidopsis and N. benthamiana, indicating that CsGAPCs negatively regulate plant immunity. Since AtGAPC knockout lines and NbGAPC knockdown plants did not exhibit any developmental defects and significantly increased plant resistance against Phytophthora infection, individual GAPC genes may be promising targets for improving disease resistance in crop plants. Further experiments are needed to investigate the effect of mutating CsGAPCs or modulating their expression in citrus by genome editing [5,48].
Las mainly colonizes within the phloem sieve cells, and infects roots before infecting leaves in citrus. Recent studies showed that papain-like cysteine proteases, which serve as targets of the SDE1 effector, are secreted into sieve elements from adjacent companion cells, while the other Las effector, SDE15, was detected in the phloem sap extract of Las-infected citrus bark tissue by western blot analysis [15,17]. Our data suggest that SDE3 binds to the cytosolic CsGAPCs in citrus, which are the housekeeping GAPDH family proteins in plants and human. Thus, it is conceivable that SDE3 is also delivered by Las into the phloem. Further studies are needed to determine how CsGAPCs suppress defense signaling and promote Las multiplication in citrus.
In animals, several pathogenic bacteria have been reported to manipulate the autophagy pathway to their own benefit, some intracellular pathogenic bacteria can be negatived by autophagy, while others (such as Shigella, Yersinia, and Listeria) can utilize autophagy to enhance their virulence [49]. In plants, autophagy, an important defense mechanism, has been shown to be involved in plant-pathogen interactions, in which pathogen effectors have evolved their abilities to modulate autophagy through distinct molecular strategies for their own benefit [50]. For instance, the P6 protein of Cauliflower mosaic virus (CaMV) inhibits autophagy and protects viral replication factory inclusions from autophagic degradation [51]. The P. syringae effector HopF3 suppresses autophagy to facilitate bacterial infection through direct interaction with ATG8 [52]. The bacterial effector SopF inhibits bacterial autophagy by preventing the V-ATPase-ATG16L1 interaction to promote Salmonella typhimurium proliferation [53]. Additionally, pathogen effectors can also induce host autophagy during plant – pathogen interactions to promote infection. The PexRD54 protein of P. infestans binds to the host autophagy protein ATG8CL to stimulate autophagosome formation, and the HopM1 protein of P. syringae activates autophagy and stimulates the autophagic removal of proteasomes to support bacterial proliferation [54,55]. It should be noted that host autophagy-related proteins can also bind to virulence factors for degradation in plants. For example, Cucumber mosaic virus (CMV) 2b protein is indirectly targeted by autophagy for degradation through its binding to the host ATG8-binding protein rgs-CaM [56,57]. In this study, we found that the expression of the Las effector SDE3 and its host target proteins CsGAPCs can suppress autophagy and also affect the stability of key autophagy components, CsATG8s, by the autophagic pathway, suggesting that the inhibition of autophagy by SDE3 and CsGAPCs might be via the CsATG8s-associated with the pathway. Taken together, these findings indicate that host autophagy facilitates cellular homeostasis via diverse strategies against various pathogens.
Although GAPC homologs have been reported to suppress autophagy in cassava (Manihot esculenta), N. benthamiana, and Arabidopsis [25,26,58], they employ diverse regulatory mechanisms to protect plants from various pathogens. For instance, NbGAPCs associate with NbATG3 to negatively regulate autophagy and promote TMV and Pst DC3000 as well as P. syringae pv. tabaci, although this depressed autophagy can also be induced by Cotton leaf curl Multan virus βC1 protein, which interferes with the GAPC-ATG3 interactions [25,36]. Our results revealed that CsGAPCs directly interact with the autophagic proteins, CsATG8s and CsATG3, but CsGAPC does not influence the stability of CsATG3, while CsGAPC expression not only abolished the nuclear-localized signal of CsATG8 but also specifically degraded CsATG8 family proteins to inhibit autophagy-mediated immunity, and this ability of CsGAPCs to degrade CsATG8s was significantly enhanced by the SDE3 effector. However, the exact mechanism of MeGAPCs and AtGAPCs in autophagy remain unknown.
Overall, the experimental data presented here reveal a complex two-layered regulatory role of autophagy in plant immunity (Figure 7E). We demonstrate that Las can subvert host autophagy to downregulate host immunity, which is achieved by the SDE3 effector. By binding to CsGAPCs, SDE3 exacerbates the degradation of CsATG8 family proteins, an autophagy component functioning in autophagosome biogenesis, thereby unveiling the role of SDE3 in the CsATG8s-mediated autophagic degradation pathway during citrus-Las interactions.
Materials and methods
Plant materials, microbial strains and growth conditions
All Arabidopsis lines and N. benthamiana plants were grown in nutrient soil in an environmentally controlled growth room at 23°C with a 16/8 h light/dark photoperiod. The transgenic and wild-type Arabidopsis lines were planted on Murashige and Skoog (MS; PhytoTech, HRY0519304A) medium at 21°C for autophagy analysis. The citrus cultivar, “Wanjincheng” orange (C. sinensis Osbeck), was used for genetic transformation. All transgenic and WT citrus plants were grown in a glasshouse at 28°C. P. capsici (Pc35) was grown in V8 medium (Campbell Soup Company, 33ZB1524) with ampicillin (50 µg/mL) at 25°C in the dark, Pst (DC3000) was cultured at LM medium (Tryptone [10 g/liter], yeast extract [6 g/liter], K2HPO4 [1.5 g/liter], NaCl [0.6 g/liter], MgSO4 [0.4 g/liter]) with streptomycin (50 µg/mL) and Xanthomonas citri subsp. citri (Xcc) was grown in Luria-Bertani (LB) medium (BactioR-tryptone [10 g/liter], BactoR-yeast extract [5 g/liter), NaCl [5 g/liter]) with kanamycin (50 µg/mL) antibiotics at 37°C.
Virus-induced virulence effector (VIVE) assay and Northern blotting
The VIVE assay was conducted as previously described [37]. Briefly, PCR products of SDE3 (without signal peptides) were ligated into the pGR106 vector under the control of the strong constitutive Cauliflower Mosaic Virus 35S promoter (CaMV 35S), which contains the entire PVX genome. The resulting construct was introduced into A. tumefaciens strain GV3101, and then infiltrated in 10-day-old leaves of N. benthamiana plants. The viral symptoms were investigated and photographed at 21 dpi. Total RNA was extracted from the newly grown leaves in infected plants, Viral RNAs were determined using northern blotting by probes against PVX coat protein (CP).
Pathogen inoculation and biomass determination assays
Pathogen inoculation assay was performed as previously described [59,60]. Briefly, approximately 3–4-week-old leaves of Arabidopsis and N. benthanmiana expressing SDE3, CsGAPC1, CsGAPC2, or CsATG8s were infected with 10–20 μL of zoospore suspension (c. 2000 zoospores) of P. capsici. Inoculated leaves were incubated in a growth chamber at 24°C for 2–3 d before analysis of disease progression. Leaves from six to 10 plants were analyzed for each treatment. The experiment was repeated twice.
Leaf discs of N. benthamiana or Arabidopsis were sampled from infection sites 2 d after P. capsici inoculation. Pure genomic DNA was extracted from leaf disks using a genomic DNA isolation kit (TIANGEN, DP350–02). Relative biomass in infected leaves was determined by the ratio of Phytophthora DNA compared with Arabidopsis or N. benthamiana DNA using qPCR analysis. Phytophthora-N. benthamiana or Arabidopsis specific primers were used (Table S3).
Mass spectrometry analysis and co-IP assay
Total protein was extracted from of N. benthamiana leaves expressing SDE3, which harbored the YFP and FLAG tags at its N and C terminus, respectively. The extracted proteins were incubated with GFP-Trap A beads (MBL, D153–11) for 3 h at 4°C. Immuno-bound proteins were denatured at 98°C and separated by SDS-PAGE. Subsequently, in-gel digestion was performed with trypsin (Promega, V5113) in 50 mM ammonium bicarbonate at 37°C overnight. The peptides were extracted with 1% (v:v) trifluoroacetic acid in 50% (v:v) acetonitrile aqueous solution.
For LC-MS/MS analysis, the extracted peptides were separated by a 60 min gradient elution at a flow rate 0.30 μL/min with a Thermo-Dionex Ultimate 3000 HPLC system. The analytical column was a fused silica capillary column (Agilent, G1364–87305). The LTQ-Orbitrap mass spectrometer (Thermo Electron Corporation, USA) was operated in the data-dependent acquisition mode using Xcalibur 2.0.7 software with a single full-scan mass spectrum in the Orbitrap followed by 20 data dependent MS/MS scans in an ion trap at 35% normalized collision energy. MS/MS spectra from each LC-MS/MS run were searched against the selected database using the Proteome Discoverer (version 1.4) searching algorithm.
Co-IP assays were carried out as described previously [61]. The PCR products were cloned into the pQBV3 or pQBV3–3*FLAG vector using a one-step cloning kit (Vazyme Biotech, C112–02-AB) and then recombined into the pEarleyGate104 or pEarleyGate100 vector. Constructs were transiently coexpressed in N. benthamiana t by Agro-infiltration. Total proteins were extracted at 48 hpi and then incubated with GFP (Abmart, M20004L), HA (KT health, KTSM1305) or FLAG (Biomed, M185–10) agarose beads at 4°C. Coprecipitation signals were detected by using an anti-FLAG (MBL, M20008S), anti-HA (MBL, M180), anti-His (MBL, D291–3), anti-MBP (NEB, E8032S) and anti-GFP (Abmart, M20004L) antibody, respectively.
BiFC and affinity-isolation assays
For BiFC assay, the full-length CDS without a stop codon of genes were PCR-amplified using Pfu polymerase, respectively (Vazyme Biotech, P505-d1-AA), the PCR products were cloned into the pQBV3 vector and then recombined into the pEarleyGate201-YN or pEarleyGate202-YC vectors [62]. The resulting constructs were transformed into the Agrobacterium GV3101 and then were transiently co-expressed in N. benthamiana leaves. Fluorescence signals were detected using an Olympus FV3000 confocal microscope (Olympus, Japan). For the affinity-isolation assay, full-length cDNA of SDE3 without the SP and CsATG8s were inserted into the pET-28a vector, CsGAPC1 and CsGAPC2 were cloned into the pMAL-c2× vector, respectively. These resulting constructs and empty vectors were expressed in E. coli strain BL21. The MBP affinity-isolation assays were conducted as described previously [63]. Briefly, MBP, MBP-CsGAPC1 and MBP-CsGAPC2 fusion proteins coupled to 50 μL MBP agarose beads (GE Healthcare, 28-9355-97) was incubated with HIS-SDE3 or HIS-CsATG8s for 3–5 h at 4°C. After washing the beads (MBL) 3–5 times with 1×phosphate buffered salt (PBS) solution (KH2PO4 [0.24 g/liter], Na2HPO4 [1.44 g/liter], NaCl [8 g/liter], KCl [0.2 g/liter], pH7.4), the co-precipitation of CsGAPC1 and CsGAPC2 with SDE3 or CsATG8s were examined by western blotting using specific antibodies (MBL).
Nuclear-cytoplasmic fractionation assay
For nuclear-cytoplasmic fractionation assays, the infiltrated leaves were grounded, mixed with 2 mL/g of lysis buffer (20 mM Tris-HCl, pH 7.5, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM sucrose [Diamond, A100335–0005], 5 mM DTT, 10 mM protease inhibitor [Sigma, P2714]) and then filtered through Miracloth (Millipore, 475855-1RCN). The obtained solution was centrifuged at 1500 × g for 10 min at 4°C. The centrifuged supernatant was centrifuged at 10,000 g for 10 min at 4°C again and the supernatant was used for cytoplasmic fraction. The centrifuged pellet was washed 4–5 times with NRBT buffer (20 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 0.2% Triton X-100 [MACKLiN, T6328-100 ML], 25% glycerol and protease inhibitor cocktail [Sigma, P8215-5 ML]) and resuspended with 500 μL of NRB2 (20 mM Tris-HCl, pH 7.5, 250 mM sucrose, 10 mM MgCl2, 0.5% Triton X-100, 5 mM β-mercaptoethanol and protease inhibitor cocktail) and slowly added 500 μL NRB3 (20 mM Tris-HCl, pH 7.5, 1.7 M sucrose, 10 mM MgCl2, 0.5% Triton X-100, 5 mM β-mercaptoethanol and protease inhibitor cocktail) on top and subjected for centrifugation at 16,000 g for 45 min. The final nuclear pellet was resuspended in lysis buffer. As quality controls for the fractionation assays, Rubisco (Abmart, M20044F) and Histone (Abmart, P30266S) were used as a cytoplasmic and a nuclear marker, respectively.
Microscopic observations and TEM analysis
For MDC staining, 10-day-roots of transgenic and WT Arabidopsis lines were soaked in MS medium and treated with 100 μM BTH (Alfa Aesar, 43730) for at least 5 h, followed by two times washing with PBS buffer and then stained with 0.05 mM MDC (Sigma,10121-91-2) in dark for 30 min before observation. MDC-incorporated structures were excited by a wavelength of 405 nm and detected at 410 to 585 nm; chloroplast autofluorescence was excited at 405 nm and detected at 635 to 708 nm.
Agrobacterium tumefaciens GV3101 harboring GFP-GAPC1and the autophagy marker CFP-NbATG8f were infiltrated into Nicotiana benthamiana leaves. About 36 h and 40 h later, 100 μM BTH used to spray the infiltrated leaves for two times. The samples were imaged using an Olympus FV3000 three-channel microscope with an excitation wavelength of 405 nm, and the emission was captured at 450 to 600 nm.
TEM observation was performed as described previously [64]. The roots of the transgenic and WT Arabidopsis lines were soaked with 100 μM BTH for 5 h at least. Leaves of sweet orange lines were infiltrated with 50 μM BTH for 24 h, ant then infiltrated with 50 μM BTH again before sampling. Thesse treated samples were fixed in 2.5% glutaraldehyde and placed at 4°C for 6 h at least, respectively. The sampled tissues treated with 100 mM PB and then post-fixed in OsO4, dehydrated in different ratio of ethanol and acetone, and then embedded indifferent ratio of resin (Structure Probe, Inc. SPI-PonTM812) and acetone. Ultrathin (100–2000 nm) sections were prepared on an ultramicrotome (LEICA EM UC7FC7) with a diamond knife and collected on Formvar-coated grids. Subsequently, sections were stained with uranyl acetate and lead citrate and examined with an electron microscope (Tecnai G2, USA).
RNA isolation, RT-PCR and Qrt-PCR
Total RNA was isolated from Arabidopsis, N. benthamiana and citrus leaves using RNAiso Plus (TaKaRa, 9109) according to the manufacturer’s protocol. For RT-PCR analysis, first-strand cDNA was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper) with the Oligo-(dT)23VN (Vazyme Biotech, R212–02-AE). qRT-PCR was used to assess the abundance of gene transcript using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech, Q711–02), and performed on a Lightcycler 480 II (Roche). Gene-coding sequences were amplified using gene specific primers (Table S1).
Transient and stable gene expression in Arabidopsis and N. benthamiana
For transient expression analysis, the validated plasmids were introduced into A. tumefaciens strain GV3101 and then infiltrated in the leaves of 3–4-week-old N. benthamiana. Protein expression was detected by western blotting at 48 hpi.
For stable expression analysis, wild-type (Col-0) Arabidopsis were used to transform via the dipping method [65]. The independent transgenic line were determined by screening for resistance and evaluated by expression of the targeting protein. The homozygous plants were used for phenotypic analysis and pathogen inoculation.
Generation of SDE3-expressing transgenic citrus and Xcc infection assay
For citrus genetic transformation, the full-length cDNA of SDE3 without the SP was inserted into a pLGN135 vector that driven by the 35S promoter [66]. The resulting construct was then introduced into the A. tumefaciens strain EHA105. Epicotyl segments from citrus seedlings were used for A. tumefaciens-mediated transformation as previously described [67]. Transgenic shoots were then identified by GUS staining as well as PCR analysis before being micrografted onto Troyer citrange seedlings in vitro. Subsequently, the resulting plantlets were further grafted onto Troyer citrange seedlings in a glasshouse. The expression of SDE3 effector in those transgenic plants was determined by RT-qPCR analysis.
Xcc infection assay was performed using in vitro pinprick inoculation and in vivo infiltration as previously described, with minor modifications [66,68]. In brief, fully mature healthy leaves of transgenic and WT citrus plants were inoculated with 1 × 108 CFU ml−1 Xcc 29–1. Citrus canker symptoms and bacterial growth were determined at 9 dpi.
‘Ca. L. asiaticus’ infection on citrus plants and HLB pathogenicity assay
Two independent lines of SDE3 transgenic citrus plants were used for HLB pathogenicity assay via a grafting method, as previously described by Clark et al. [15]. Wild-type sweet orange cultivar “Wanjincheng” plants of the same age were used as controls. Each SDE3 transgenic line represents an independent transgenic event. For the “Ca. L. asiaticus” graft inoculation, buds from “Ca. L. asiaticus”-infected greenhouse-grown sweet orange plants, with obvious disease symptoms and the presence of “Ca. L. asiaticus” confirmed by qPCR, were grafted onto SDE3 transgenic plants with two buds for each branch. Leaf samples were collected from the graft-inoculated grapefruit plants monthly beginning immediately after grafting until one month postgrafting. Midribs of three mature leaves were collected for each branch, which were individually processed for DNA extraction and “Ca. L. asiaticus” quantification. qPCR was performed using gene-specific primers (Table S1). The Ct value of each amplicon represents the “Ca. L. asiaticus” genomic copy numbers in 100 ng of DNA.
Phylogenetic analyses
The deduced protein sequences of SDE3 homologs in bacteria and GAPDH family proteins in plants were downloaded by the FungiDB and NCBI Blastp search program. The amino acid sequences were aligned using Clustal Omega and implemented in BoxShade server. The phylogenetic tree was produced using the Neighbor-Joining method in MEGA 7 [69].
Statistical analyses
All statistical analyses were performed with the Duncan’s multiple range test.
Acknowledgements
We thank Prof. Yule Liu (Tsinghua University) for providing the CFP-NbATG8 plasmid. Prof. Feng Yu (Hunan University) for providing Arabidopsis mutants (atgapc1 and atgapc2).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (32172359, 32072502, 32001883, and 31871925), the National Key Research and Development Program of China (2018YFD0201500, 2021YFD1400800), the Science and Technology Commission of Shanghai Municipality (18DZ2260500).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].JM B. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 2006;88:7–37. [Google Scholar]
- [2].Blaustein RA, Lorca GL, Teplitski M. Challenges for managing Candidatus Liberibacter spp. (Huanglongbing disease pathogen): current control measures and future directions. Phytopathology. 2018;108:424–435. [DOI] [PubMed] [Google Scholar]
- [3].Hu B, Rao MJ, Deng X, et al. Molecular signatures between citrus and Candidatus Liberibacter asiaticus. PLOS Pathog. 2021;17:e1010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang N. A perspective of citrus Huanglongbing in the context of the Mediterranean Basin. J Plant Pathol. 2020;102:635–640. [Google Scholar]
- [5].Wang N, Pierson EA, Setubal JC, et al. The Candidatus Liberibacter-host interface: insights into pathogenesis mechanisms and disease control. Annu Rev Phytopathol. 2017;55:451–482. [DOI] [PubMed] [Google Scholar]
- [6].Barnett MJ, Solow-Cordero DE, Long SR. A high-throughput system to identify inhibitors of Candidatus Liberibacter asiaticus transcription regulators. Proc Natl Acad Sci, USA. 2019;116:18009–18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang CY, Araujo K, Sanchez JN, et al. A stable antimicrobial peptide with dual functions of treating and preventing citrus Huanglongbing. Proc Natl Acad Sci, USA. 2021;118(6):e2019628118. DOI: 10.1073/pnas.2019628118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Duan Y, Zhou L, Hall DG, et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact. 2009;22:1011–1020. [DOI] [PubMed] [Google Scholar]
- [9].Shi Q, Pitino M, Zhang S, et al. Temporal and spatial detection of Candidatus Liberibacter asiaticus putative effector transcripts during interaction with Huanglongbing-susceptible, -tolerant, and -resistant citrus hosts. BMC Plant Biol. 2019;19:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prasad S, Xu J, Zhang YZ, et al. SEC-translocon dependent extracytoplasmic proteins of Candidatus Liberibacter asiaticus. Front Microbiol. 2016;7:1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yan Q, Sreedharan A, Wei S, et al. Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol Plant Pathol. 2013;14:391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pagliaccia D, Shi JX, Pang ZQ, et al. A pathogen secreted protein as a detection marker for citrus huanglongbing. Front Microbiol. 2017;8:2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pitino M, Armstrong CM, Cano LM, et al. Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front Plant Sci. 2016;7:982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tomkins M, Kliot A, Maree AFM, et al. A multi-layered mechanistic modelling approach to understand how effector genes extend beyond phytoplasma to modulate plant hosts, insect vectors and the environment. Curr Opin Plant Biol. 2018;44:39–48. [DOI] [PubMed] [Google Scholar]
- [15].Clark K, Franco JY, Schwizer S, et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat Commun. 2018;9:1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Clark KJ, Pang Z, Trinh J, et al. Sec-delivered effector 1 (SDE1) of ‘Candidatus Liberibacter asiaticus’ promotes citrus huanglongbing. Mol Plant Microbe Interact. 2020;33:1394–1404. [DOI] [PubMed] [Google Scholar]
- [17].Pang ZQ, Zhang L, Coaker G, et al. Citrus CsACD2 is a target of Candidatus Liberibacter Asiaticus in huanglongbing disease. Plant Physiol. 2020;184:792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Du PX, Zhang C, Zou XP, et al. “Candidatus Liberibacter asiaticus” secretes nonclassically secreted proteins that suppress host hypersensitive cell death and induce expression of plant pathogenesis-related proteins. Appl Environ Microb. 2021;87(8):e00019–21. DOI: 10.1128/AEM.00019-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang C, Du PX, Yan HL, et al. A sec-dependent secretory protein of the huanglongbing-associated pathogen suppresses hypersensitive cell death in Nicotiana benthamiana. Front Microbiol. 2020;11:594669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang C, Wang XF, Liu XL, et al. A novel ‘Candidatus Liberibacter asiaticus’-encoded sec-dependent secretory protein suppresses programmed cell death in Nicotiana benthamiana. Int J Mol Sci. 2019;20(22):5802. DOI: 10.3390/ijms20225802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Plaxton WC. The organization and regulation of plant glycolysis. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:185–214. [DOI] [PubMed] [Google Scholar]
- [22].Petersen J, Brinkmann H, Cerff R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol. 2003;57:16–26. [DOI] [PubMed] [Google Scholar]
- [23].Li TT, Liu MX, Feng X, et al. Glyceraldehyde-3-phosphate dehydrogenase is activated by lysine 254 acetylation in response to glucose signal. J Biol Chem. 2014;289:3775–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Anoman AD, Munoz-Bertomeu J, Rosa-Tellez S, et al. Plastidial glycolytic glyceraldehyde-3-phosphate dehydrogenase is an important determinant in the carbon and nitrogen metabolism of heterotrophic cells in Arabidopsis. Plant Physiol. 2015;169:1619–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han S, Wang Y, Zheng X, et al. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell. 2015;27:1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Henry E, Fung N, Liu J, et al. Beyond glycolysis: gAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015;11:e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Garcin ED. GAPDH as a model non-canonical AU-rich RNA binding protein. Semin Cell Dev Biol. 2019;86:162–173. [DOI] [PubMed] [Google Scholar]
- [28].Howard TP, Lloyd JC, Raines CA. Inter-species variation in the oligomeric states of the higher plant Calvin cycle enzymes glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. J Exp Bot. 2011;62:3799–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schneider M, Knuesting J, Birkholz O, et al. Cytosolic GAPDH as a redox-dependent regulator of energy metabolism. BMC Plant Biol. 2018;18(1):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Miao LK, Chen CL, Yao L, et al. Genome-wide identification, characterization, interaction network and expression profile of GAPDH gene family in sweet orange (Citrus sinensis). Peer J. 2019;7:e7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sirover MA. Moonlighting glyceraldehyde-3-phosphate dehydrogenase: posttranslational modification, protein and nucleic acid interactions in normal cells and in human pathology. Crit Rev Biochem Mol. 2020;55:354–371. [DOI] [PubMed] [Google Scholar]
- [32].Zaffagnini M, Fermani S, Costa A, et al. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013;4:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vescovi M, Zaffagnini M, Festa M, et al. Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 2013;162:333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Baek D, Jin Y, Jeong JC, et al. Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry. 2008;69:333–338. [DOI] [PubMed] [Google Scholar]
- [35].Guo L, Devaiah SP, Narasimhan R, et al. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Ddelta to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell. 2012;24:2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ismayil A, Yang M, Haxim Y, et al. Cotton leaf curl Multan virus beta C1 protein induces autophagy by disrupting the interaction of autophagy-related protein 3 with glyceraldehyde-3-phosphate dehydrogenases. Plant Cell. 2020;32:1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shi JX, Zhu YH, Li M, et al. Establishment of a novel virus-induced virulence effector assay for the identification of virulence effectors of plant pathogens using a PVX-based expression vector. Mol Plant Pathol. 2020;21:1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Mol Plant Pathol. 2003;4:141–157. [DOI] [PubMed] [Google Scholar]
- [39].Xu G, Wang S, Han S, et al. Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy. 2017;13:1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang M, Zhang YL, Xie XL, et al. Barley stripe mosaic virus gamma b protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell. 2018;30:1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang Y, Yu B, Zhao J, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013;25:1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010;62:483–493. [DOI] [PubMed] [Google Scholar]
- [43].Suttangkakul A, Li FQ, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bu F, Yang M, Guo X, et al. Multiple functions of ATG8 family proteins in plant autophagy. Front Cell Dev Biol. 2020;8:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Graham J, Gottwald T, Setamou M. Status of Huanglongbing (HLB) outbreaks in Florida, California and Texas. Trop Plant Pathol. 2020;45:265–278. [Google Scholar]
- [46].Testard A, Da Silva D, Ormancey M, et al. Calcium- and nitric oxide-dependent nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in response to long chain bases in tobacco BY-2 cells. Plant Cell Physiol. 2016;57:2221–2231. [DOI] [PubMed] [Google Scholar]
- [47].Truong NM, Chen Y, Mejias J, et al. The Meloidogyne incognita nuclear effector MiEFF1 interacts with Arabidopsis cytosolic glyceraldehyde-3-phosphate dehydrogenases to promote parasitism. Front Plant Sci. 2021;12:641480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jia H, Zhang Y, Orbovic V, et al. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J. 2017;15:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Leary AY, Savage Z, Tumtas Y, et al. Contrasting and emerging roles of autophagy in plant immunity. Curr Opin Plant Biol. 2019;52:46–53. [DOI] [PubMed] [Google Scholar]
- [51].Hafren A, Macia JL, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci, USA. 2017;114:E2026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lal NK, Thanasuwat B, Huang PJ, et al. Phytopathogen effectors use multiple mechanisms to manipulate plant autophagy. Cell Host Microbe. 2020;28:558–571. [DOI] [PubMed] [Google Scholar]
- [53].Xu Y, Zhou P, Cheng S, et al. A Bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 2019;178:552–566.e20. [DOI] [PubMed] [Google Scholar]
- [54].Dagdas YF, Belhaj K, Maqbool A, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife. 2016;5:e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ustun S, Hafren A, Liu Q, et al. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell. 2018;30:668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nakahara KS, Masuta C, Yamada S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci, USA. 2012;109:10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shukla A, Hoffmann G, Kushwaha NK, et al. Salicylic acid and the viral virulence factor 2b regulate the divergent roles of autophagy during cucumber mosaic virus infection. Autophagy. 2022;18:1450–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zeng H, Xie Y, Liu G, et al. Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Mol Biol. 2018;97:201–214. [DOI] [PubMed] [Google Scholar]
- [59].Zhang P, Jia Y, Shi J, et al. The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytol. 2019;223:839–852. [DOI] [PubMed] [Google Scholar]
- [60].Gui X, Zhang P, Wang D, et al. Phytophthora effector PSR1 hijacks the host pre-Mrna splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell. 2022;34(9):3443–3459. DOI: 10.1093/plcell/koac176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Qiao Y, Shi J, Zhai Y, et al. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci, USA. 2015;112:5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lu Q, Tang X, Tian G, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. [DOI] [PubMed] [Google Scholar]
- [63].Chen C, He B, Liu X, et al. Pyrophosphate-fructose 6-phosphate 1-phosphotransferase (PFP1) regulates starch biosynthesis and seed development via heterotetramer formation in rice (Oryza sativa.L). Plant Biotechnol J. 2020;18:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li F, Zhao N, Li Z, et al. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLOS Pathog. 2017;13:e1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. [DOI] [PubMed] [Google Scholar]
- [66].Zou XP, Du MX, Liu YN, et al. CsLOB1 regulates susceptibility to citrus canker through promoting cell proliferation in citrus. Plant J. 2021;106:1039–1057. [DOI] [PubMed] [Google Scholar]
- [67].Peng AH, Xu LZ, He YR, et al. Efficient production of marker-free transgenic ‘Tarocco’ blood orange (Citrus sinensis Osbeck) with enhanced resistance to citrus canker using a Cre/loxP site-recombination system. Plant Cell Tiss Org. 2015;123:1–13. [Google Scholar]
- [68].Jia HG, Wang N. Generation of homozygous canker-resistant citrus in the T0 generation using CRISPR-SpCas9p. Plant Biotechnol J. 2020;18:1990–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]


