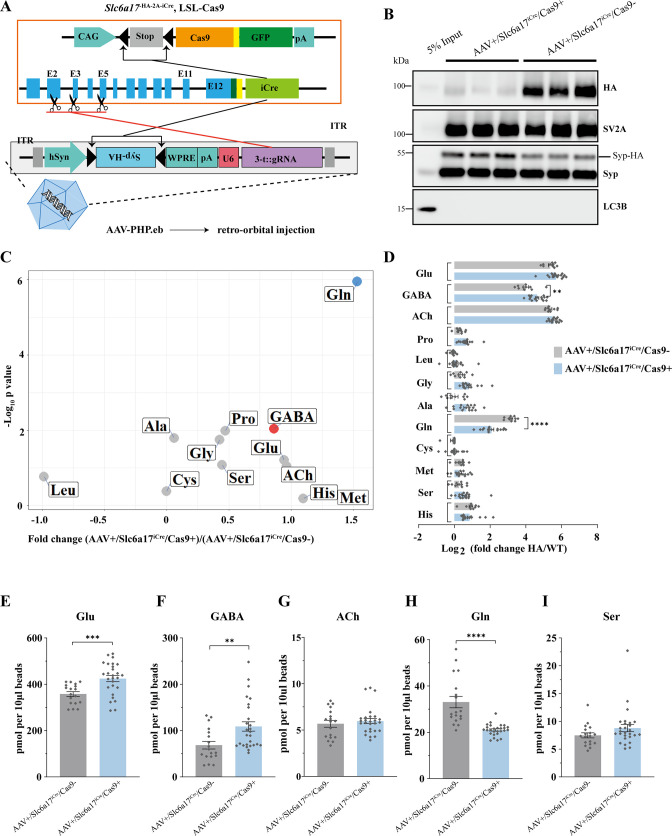
Figure 9. Physiological requirement of SLC6A17 for glutamine (Gln) transport into synaptic vesicles (SVs) in vivo.
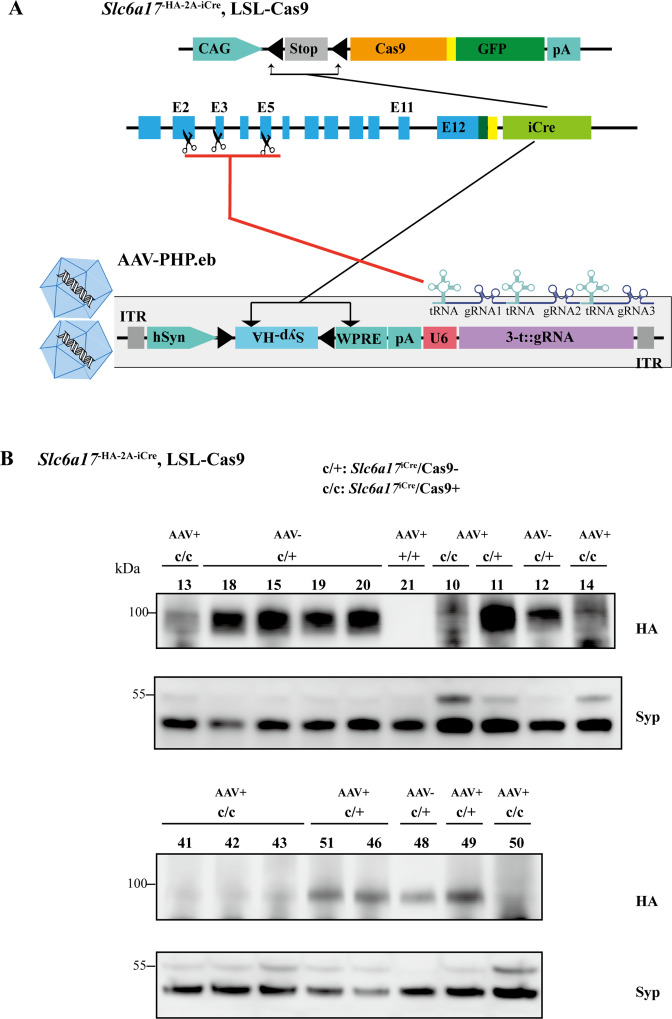
(A) A schematic diagram illustrating the strategy of Cas9-mediated cleavage of Slc6a17 specifically in Slc6a17-positive neurons, and simultaneous labeling of all SVs in these neurons by Syp-HA. (B) Immunoblot results showing SLC6A17 protein was significantly reduced in targeted neurons, while Syp-HA was efficiently tagged onto the SVs of these neurons. (C) Volcano plot of contents of SVs from AAV-sgRNA/Slc6a17iCre/Cas9+ targeted neurons compared to contents of SVs from control (AAV-sgRNA /Slc6a17iCre/Cas9-) neurons. Glu, GABA, ACh, and the nine previously reported substrates of SLC6A17 are listed. (D) Ratios of the level of a molecule from the SVs of AAV-sgRNA /Slc6a17iCre/Cas9+ neurons vs. the level of the same molecule from SVs of AAV-sgRNA/Slc6a17iCre/Cas9- neurons shown as fold change (log2 transformed). GABA level was significantly increased (p=0.0034 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-). Gln level was significantly decreased (p<0.0001 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-). (E–H) Contents of SVs from Slc6a17 containing neurons were quantified to mole per 10 μl HA beads (n = 18, 27 for Slc6a17iCre/Cas9- and Slc6a17iCre/Cas9+, respectively, from six and nine different animals with three replicates each): Glu (E, p=0.0005 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-); GABA (F, p=0.0032 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-); Gln (G, p<0.0001 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-); Ser (H, p=0.0979 for AAV-sgRNA /Slc6a17iCre/Cas9+ vs. AAV-sgRNA /Slc6a17iCre/Cas9-).