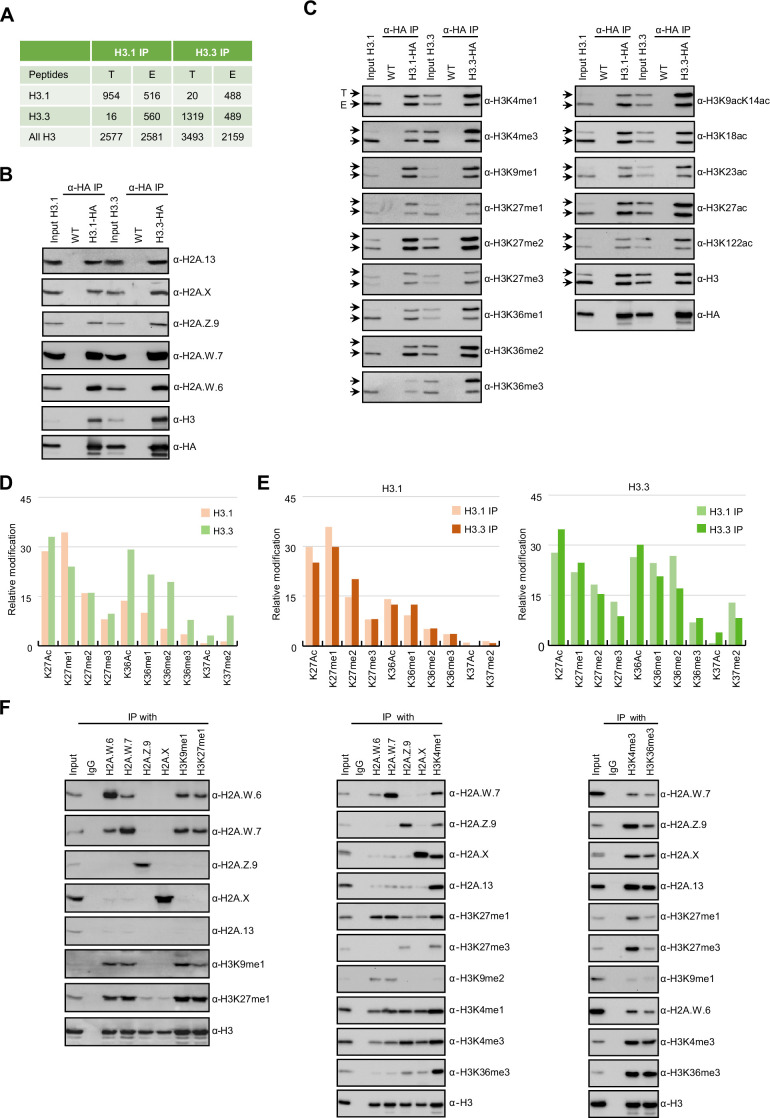
Figure 1. Biochemical analysis of the association between histone variants and histone marks.
(A) Histones H3.1 and H3.3 form homotypic and heterotypic nucleosomes. Spectral counts of H3.1- and H3.3-specific peptides in the respective immunoprecipitations (T – transgenic, E – endogenous H3.1 and H3.3). (B) H2A variants do not preferentially associate with H3.1- or H3.3-containing nucleosomes. HA-tagged H3.1 and H3.3 mononucleosomes were immunoprecipitated with HA agarose and analyzed for the presence of H2A variants by immunoblotting. (C) Histone H3 marks are present on both H3.1 and H3.3. HA-tagged H3.1 and H3.3 mononucleosomes were immunoprecipitated with HA agarose and analyzed for the presence of H3 marks by immunoblotting. Arrows indicate transgenic (T) and endogenous (E) H3. (D) Mass spectrometry (MS) analysis of cumulative H3K27, H3K36, and H3K37 modifications on H3.1 and H3.3. All measured spectra corresponding to H3.1 and H3.3 peptides from both IPs were used for analysis. (E) Relative abundance of H3K27, H3K36, and H3K37 modifications on H3.1 variant analyzed separately from MS data of H3.1 and H3.3 purified nucleosomes (left panel). Relative abundance of H3K27, H3K36, and H3K37 modifications on H3.3 variant analyzed separately from MS data of H3.1 and H3.3 purified nucleosomes (right panel). (F) Co-occurrence of H2A variants and H3 marks. Mononucleosomes were immunoprecipitated with the indicated antibodies and analyzed for the presence of H2A variants and H3 marks by western blotting. Original pictures of the gels are provided in Figure 1—source data 1, Figure 1—source data 2 and Figure 1—source data 3.