Figure 6.
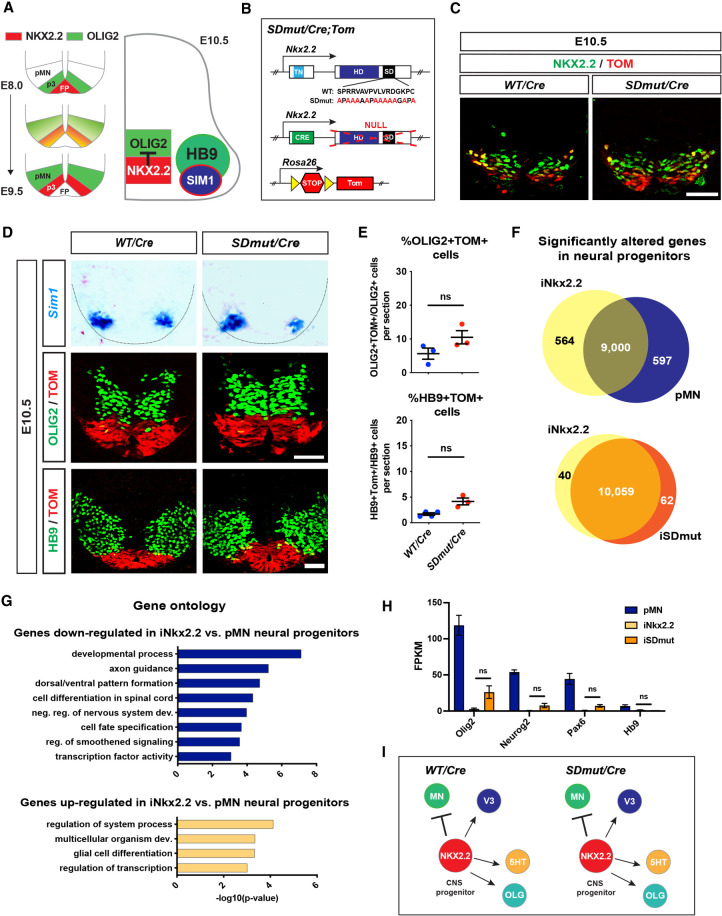
NKX2.2-dependent CNS populations are not affected by mutation of the SD. (A) Schematic representing the onset of NKX2.2-mediated repression of motor neuron (MN) identity. (B) Diagram of NKX2.2 alleles used to generate Nkx2.2SDmut/Cre (SDmut/Cre) and Nkx2.2Cre/+ (WT/Cre) mice. The Rosa26-Tomato reporter allele (TOM) was included to enable lineage tracing. (C) NKX2.2 and TOM expression in E10.5 embryos. (D) In situ hybridization of Sim1, as well as OLIG2 and HB9 immunostaining, in Nkx2.2SDmut/Cre and Nkx2.2Cre/+ E10.5 spinal cords. (E) Quantification of the percentage of OLIG2+TOM+ and HB9+TOM+ cells from D. (F) RNA-seq analysis of mouse embryonic stem cells (ESCs) differentiated into control motor neuron progenitors (pMN) and pMN progenitors in which either wild-type (iNKX2.2) or SD mutant (iSDmut) NKX2.2 was ectopically induced. (G) Gene ontology (GO) analysis of transcripts differentially expressed in iNKX2.2 progenitors compared with pMN cells. (H) FPKM values of TFs necessary for MN development. (I) Schematic showing that all examined NKX2.2-dependent CNS populations (see Supplemental Fig. S8) are not affected by mutation of the SD. Data are presented as mean ± SEM. (ns) Not significant. Scale bars, 50 µm.