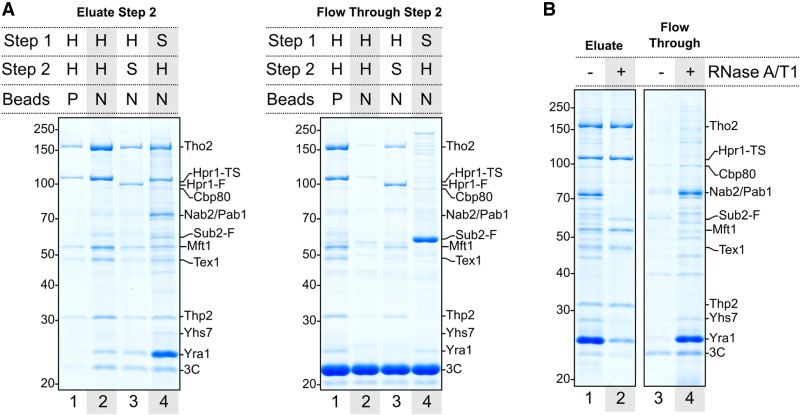
Figure 1.
Isolation of S. cerevisiae nuclear mRNPs. (A) Coomassie-stained 10% SDS-PAGE gels of SDS eluates (left) or TCA-precipitated unbound fractions (right) from the final purification step. The first affinity capture (step 1, via protein A) was always performed using nonporous magnetic beads. Elution in the first step was carried out with 3C protease. The second affinity capture (step 2, Twin-Strep) was performed using either porous (P) or nonporous (N) magnetic beads. H (Hpr1) and S (Sub2) indicate the tagged protein used as bait in the corresponding affinity capture step. Band annotations are based on mass spectrometry fingerprinting (Supplemental Fig. S1) and include the presence of tags remaining after elution. (F) FLAG, (TS) Twin-Strep. (B) Coomassie-stained 10% SDS-PAGE gels of SDS eluates (left) or TCA-precipitated unbound fractions (right) with or without RNase A/T1 mix treatment on beads prior to elution from the second affinity step.