Figure 5.
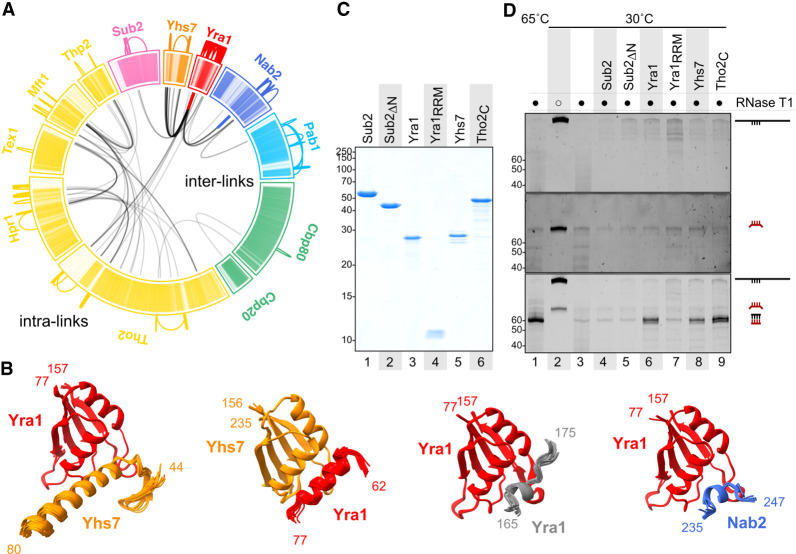
Protein and RNA interaction networks in nuclear mRNPs. (A) Location of BS3 cross-links in native particles as identified by mass spectrometry. Links between peptides within the same protein (intralinks) are shown in color on the outer region. Links between peptides originating from different proteins (interlinks) are shown in black inside the circle. Inside the circle, colored interlinks represent cross-linking between two copies of the same protein. The sequence of each protein is schematized in individual sectors and colored per protein type (as in Fig. 3C,E). In each protein sector, each line represents a residue (N terminus to C terminus, clockwise) with color intensities corresponding to scaled pLDDT values (AlphaFold per-residue confidence metric for individual predictions). Low pLDDT (which correlates with disorder) is represented in lighter shades. Darker shades (higher pLDDT) correlate well with folded regions. (B) AlphaFold predictions of protein fragments selected around identified cross-links in A. Twenty-five models predicted with AlphaFold were aligned on the RRM domains of the proteins indicated. Only domains that converged to similar structural features are displayed. Shown are 19 out of 25 models for Yra1_RRM–Yhs7, 20 out of 25 models for Yhs7_RRM–Yra1, 25 out of 25 models for Yra1_RRM–Yra1, and 15 out of 25 models for Yra1_RRM–Nab2. (C) Purified recombinant proteins separated on a 15% Coomassie-stained SDS-PAGE used as input in an RNA-annealing assay (shown in D). (D) RNA-annealing assay. A 315-nt in vitro transcribed RNA substrate derived from HHF1 mRNA (in black) was mixed with an 80-nt probe (in red). The probe contained a 60-nt sequence of perfect complementarity to part of the substrate flanked by 10-nt nonpairing extensions on either side. After incubation at the indicated temperatures with the corresponding proteins, treatment with RNase T1 digested all unpaired fragments. The remaining RNA was separated on a 14% urea-PAGE and stained with SYBR Gold. (Top) Substrate alone. (Middle) Probe alone. (Bottom) Both substrate and probe present.