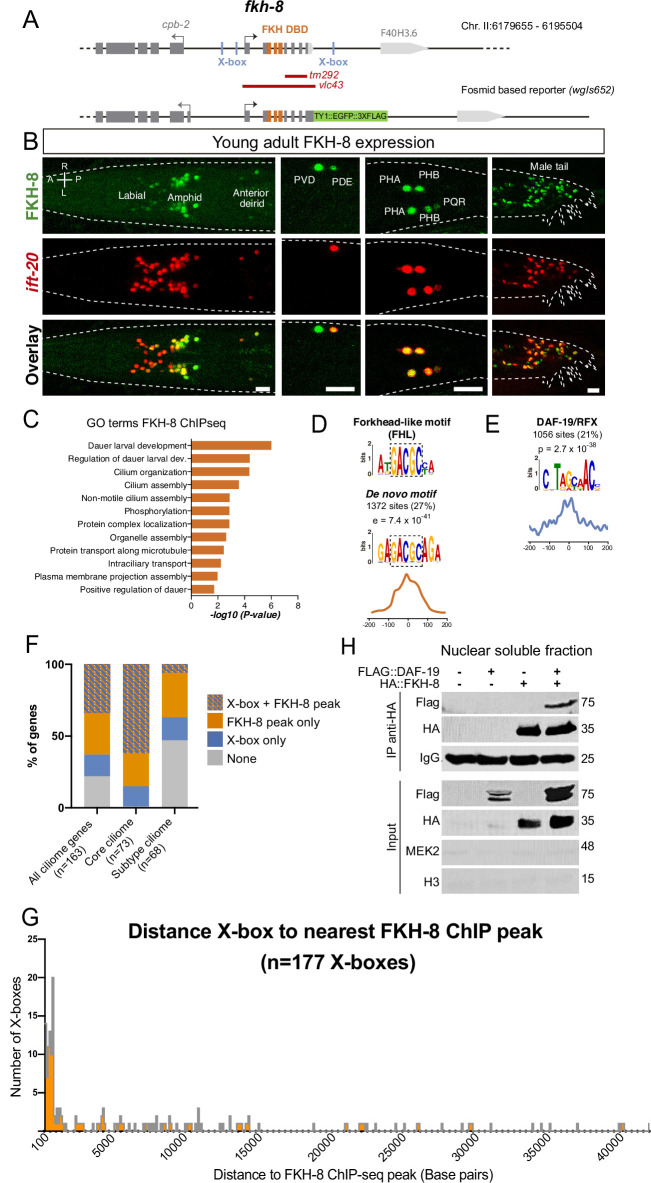
Figure 2. FKH-8 is expressed in sensory ciliated neurons, binds ciliome genes near DAF-19 X-boxes and physically interacts with DAF-19.
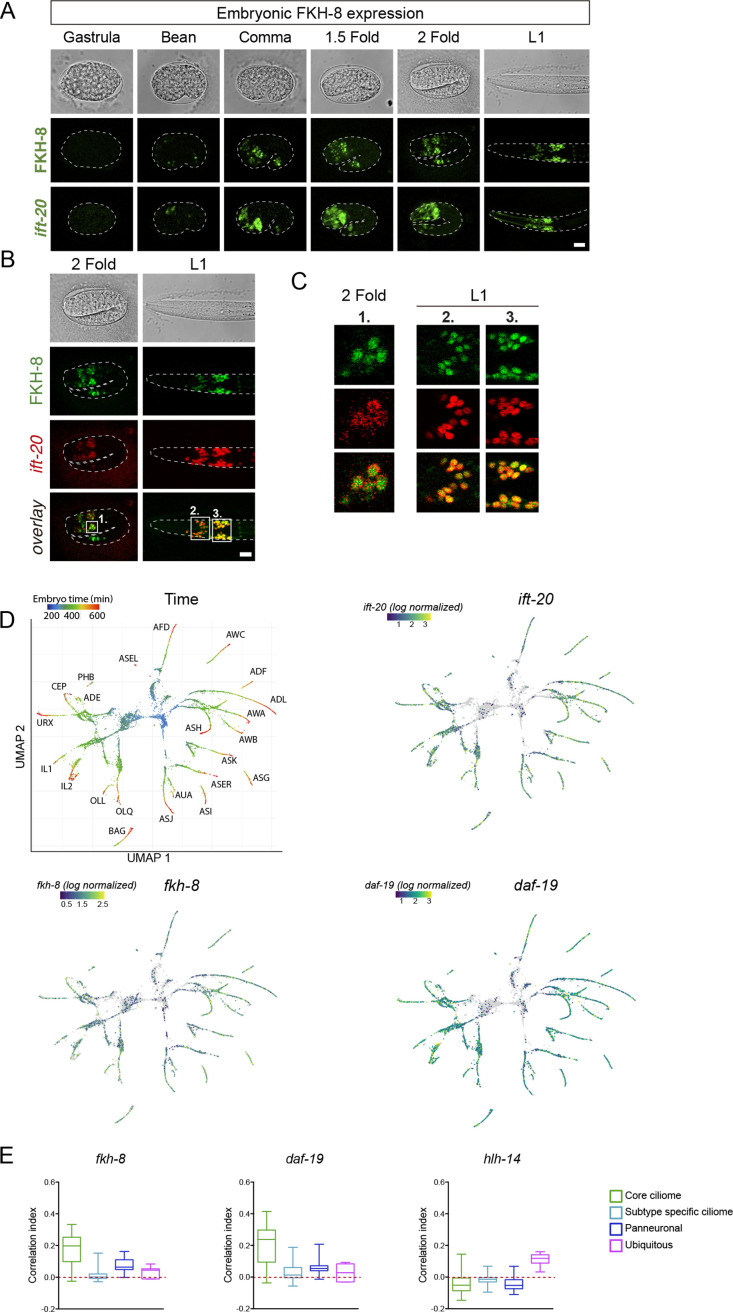
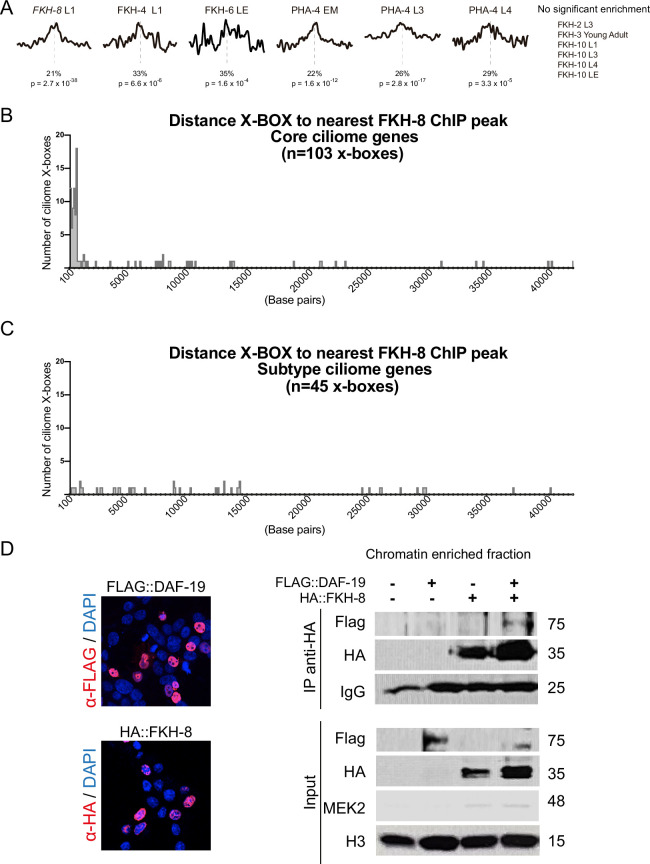
(A) fkh-8 locus (top) and fosmid based fkh-8 reporter (bottom). Grey boxes represent exons and orange boxes correspond to exons coding for the FKH DNA binding domain (DBD). Putative daf-19/RFX binding sites (X-boxes) are depicted with blue lines. Red bars indicate extension for the corresponding deletion alleles. (B) Dorso-ventral views of young adult animals expressing both the fosmid-based FKH-8::GFP reporter (in green) and an integrated reporter for the panciliary marker ift-20 (in red). A: anterior, P: posterior, R: right, L: left. Scale bar = 10 µm. See Figure 2—source data 1 for quantification and Figure 2—figure supplement 1 for embryonic expression patterns and expression correlation with DAF-19 and ciliome genes. (C) Genes associated to nearby FKH-8 binding events enrich Gene Ontology terms related to cilia regulated processes and/or functions. Data correspond to adjusted p-value. See Figure 2—source data 2 for gene lists associated to GO terms (D) De novo motif analysis of FKH-8 ChIP-seq data identifies a motif present in 27% of peaks, enriched at central positions, that matches a Forkhead like (FHL) motif. (E) DAF-19/RFX binding motifs (PWM M1534_1.02) are present in 21% of the FKH-8 bound regions and are enriched at central positions. See Figure 2—figure supplement 2 for similar analysis on additional FKH ChIP-seq data sets. (F) Distribution of ciliome genes in four different categories: (1) genes with both X-box motifs and FKH-8 binding events; (2) genes with only FKH-8 binding; (3) Genes with X-box motifs only and (4) Genes with neither FKH-8 binding or X-boxes. Most ciliome genes contain both X-boxes and FKH-8 peaks, this dual signature is highly prevalent in core ciliome genes while is minoritary in subtype ciliome genes. See Figure 2—source data 2 for gene lists associated to each signature. (G) Distance between X-boxes found in ciliome genes and the center of the nearest FKH-8 ChIP-seq peak. 42% of X-boxes are located less than 600 bp from a FKH-8 ChIP-seq peak. See Figure 2—figure supplement 2 for differential analysis of core and subtype ciliome genes. (H) Co-immuno precipitation of HA tag FKH-8 and FLAG tag DAF-19 expressed in HEK293 cells shows physical interaction between both transcription factors in the soluble fraction of nuclear extracts. MEK2 is used to assess for the presence of cytoplasmic components and Histone H3 to assess the presence of chromatin. See Figure 2—source data 3 for original blots and Figure 2—figure supplement 2 for additional analysis of interaction in chromatin associated fractions.