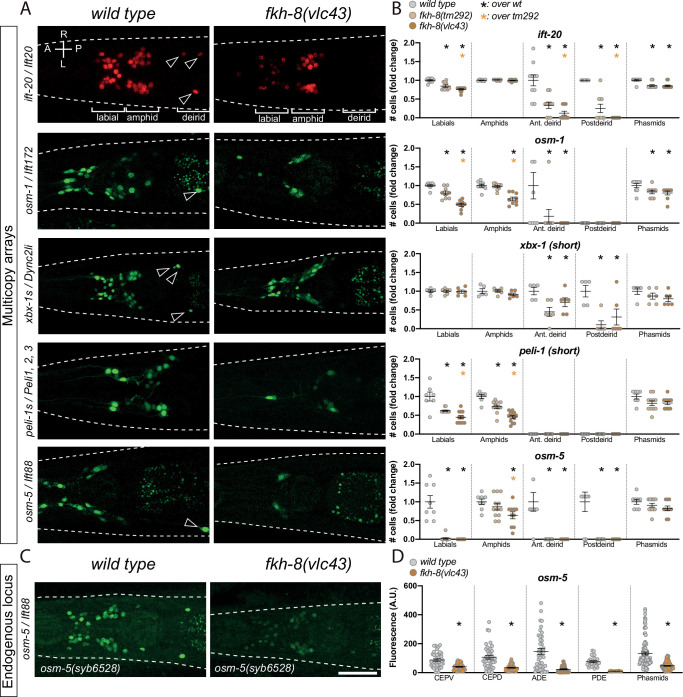
Figure 3. FKH-8 TF and FKH-binding sites are required for correct core ciliome gene reporter expression.
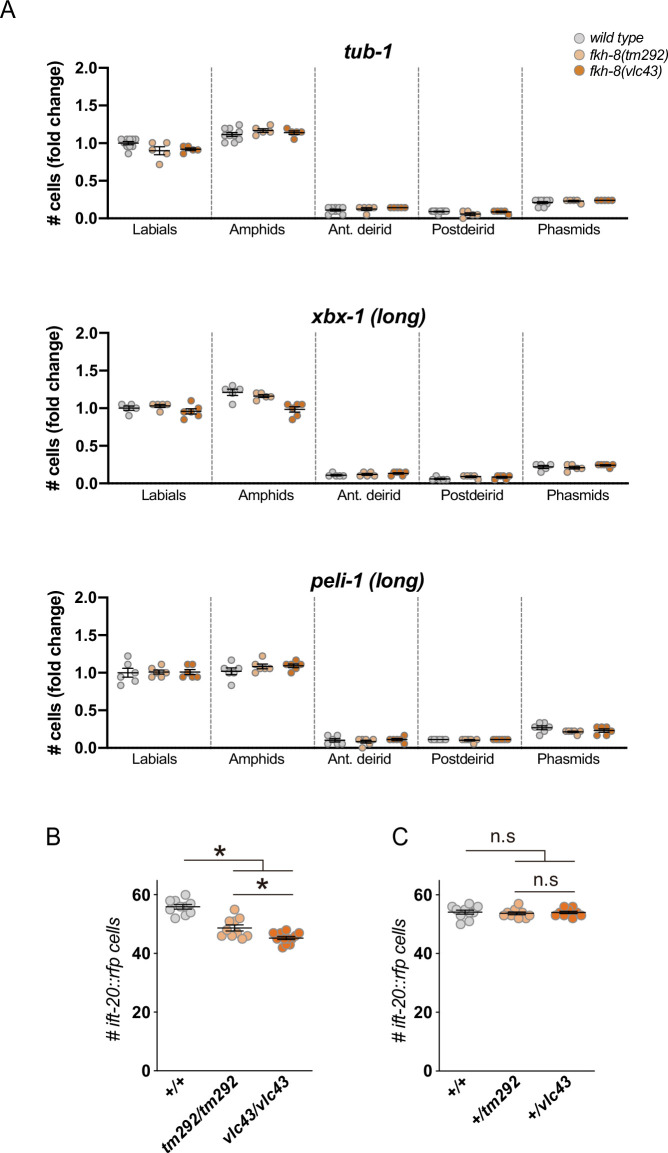
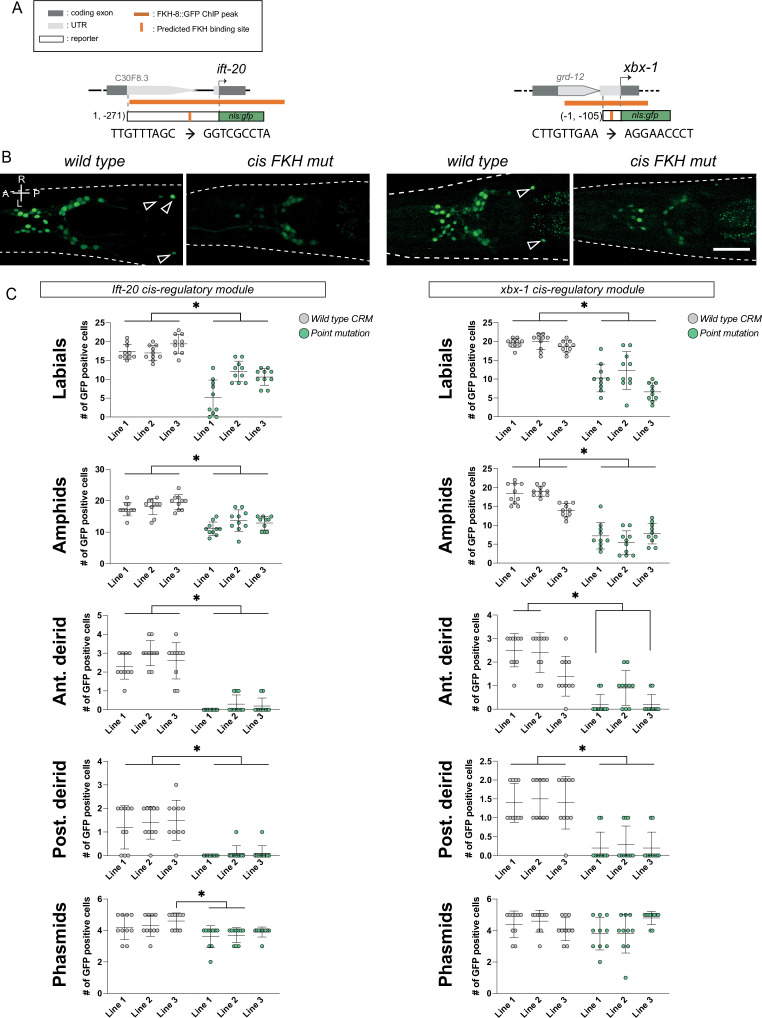
(A) Dorso-ventral images from young adult heads expressing different core ciliome multicopy array gene reporters in wild type and fkh-8(vlc43) null mutant animals. All reporters are extrachromosomal arrays except for ift-20 reporter which is integrated. Arrow heads point deirid expression lost in the mutant. A: anterior, P: posterior, R: right, L: left. (B) Quantification of the number of gfp-positive cells in five distinct anatomical regions for each reporter in wild type, fkh-8(tm292) hypomorphic allele and fkh-8(vlc43) null mutant. To facilitate comparisons, values in each region are normalized to controls. The same extrachromosomal line was analyzed in the different genetic backgrounds. Each dot represents the number of reporter-expressing neurons scored in a single animal. Mean and standard error are represented. Black asterisk denotes significantly different from wild type and orange asterisk indicates vlc43 is significantly different from tm292 allele. Sample sizes for each genetic background: ift-20: n≥10; osm-9: n=9; osm-5: n≥8; peli-1: n≥7; xbx-1: n≥5. See Figure 3—source data 1 for raw scoring data, Figure 3—figure supplement 1 for analysis of the hypomorphic recessive nature of the tm292 allele and quantification of additional reporters not affected in fkh-8 mutants, see Figure 3—figure supplement 2 and Figure 3—source data 2 for functional characterization of predicted FKH binding sites in ift-20 and xbx-1 regulatory regions. (C) Dorso-ventral images from young adult heads expressing GFP from the endogenously tagged osm-5 locus [osm-5(syb6528), osm-5::SL2::GFP::H2B] in wild type and fkh-8(vlc43) null mutant. A global decrease in fluorescence intensity is detected in fkh-8(vlc43) animals compared to wild type. Scale bar = 25 µm. (D) Fluorescence intensity level quantification in specific ciliated neuron populations shows significant reduction of expression in fkh-8(vlc43) animals. A. U.: arbitrary units. See Figure 3—source data 1 for raw scoring data. n≥20 for each cell type and genetic background.