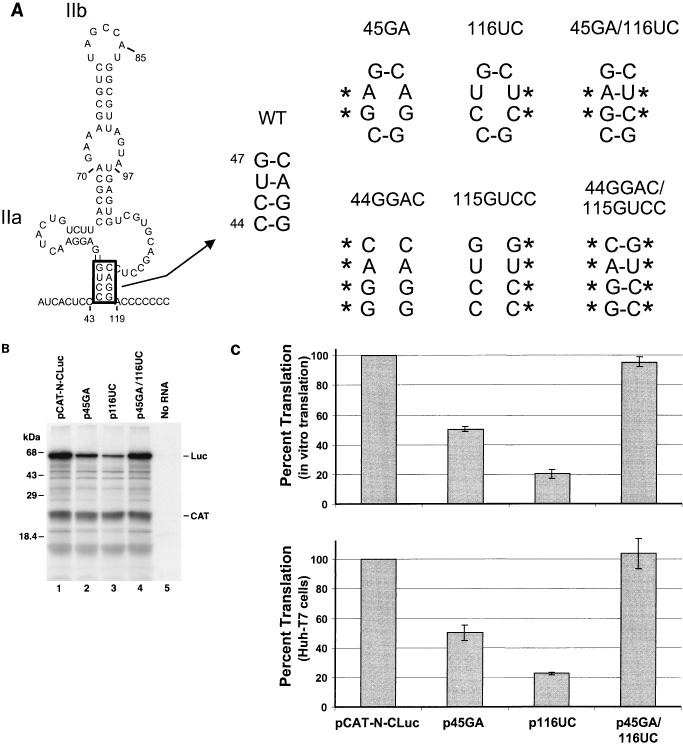
FIG. 2.
Mutational analysis of the putative helical segment at the base of domain II that is common to the two structures shown in Fig. 1. (A) Mutant plasmid p45GA contains a CU- to GA-substitution at nt 45 and 46, while p116UC contains an AG-to-UC substitution within the predicted complementary strand at nt 116 and 117. Both mutations destabilize the helix at the base of domain II. Plasmid p45GA/116UC contains both of these mutations, potentially allowing restoration of the helix. Also shown are similar mutants containing 4-nt insertions (p44GGAC, etc.). WT, wild type. (B) SDS-PAGE of translation products from coupled transcription-translation reactions programmed with plasmids pCAT-N-CLuc (wild-type; lane 1), p45GA (lane 2), p116UC (lane 3), and p45GA/116UC (lane 4). No RNA, negative transcription control. Luc, core-luciferase fusion protein. (C) Mean relative IRES activities of mutants, measured as the ratio of luciferase to CAT activity produced in vitro in reticulocyte lysate (top) or in vivo in vTF7-3 infected Huh-T7 cells (bottom). The ratio of luciferase to CAT activity obtained with pCAT-N-CLuc was accorded a value of 100%. Error bars denote standard errors of values obtained in replicate experiments.