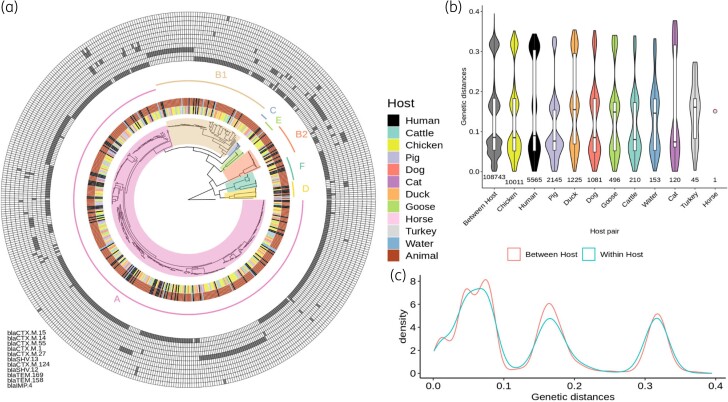
Figure 3.
Global phylogeny of 510 ESBL-Ec and associated metrics. (a) Phylogenetic maximum likelihood tree of 510 ESBL-Ec genomes built from 179 365 core and non-recombinant SNPs. Hosts for each isolate and compartments (human, animal, water) are indicated in the two most central internal rings, respectively, whereas inferred phylogroups are designed both on the phylogenetic tree and on the next ring. A matrix of the presence of the most common resistance genes (prevalence >1%) is depicted in the outer rings, with the names of the genes listed from the inner to the outer lines in the matrix. (b) Comparison of pairwise phylogenetic distances computed between strains within each host. (c) Comparison of pairwise phylogenetic distances computed between versus within hosts. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.