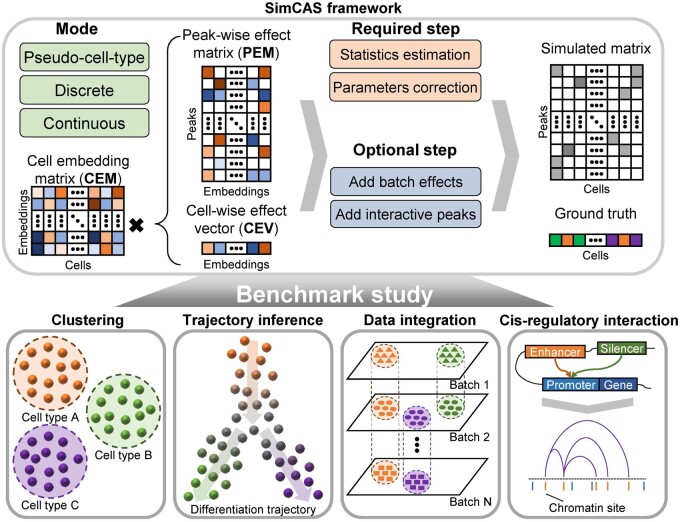
Figure 1.
A graphical illustration of simCAS framework. simCAS provides three modes to simulate cells with different states: pseudo-cell-type mode, discrete mode, and continuous mode. simCAS first generate cell- and peak-wise low-dimensional embeddings with user-defined ground truths. Then, the distributions of statistics in real data are estimated for correcting the parameters. A peak-by-cell matrix is finally generated from the corrected parameters. The batch effects and interaction peaks can be added optionally during the simulation in different modes. The synthetic data generated by simCAS can be used to benchmark four major computational tasks of analyzing scCAS data: cell clustering, trajectory inference, data integration, and cis-regulatory interaction inference.