Fig. 3.
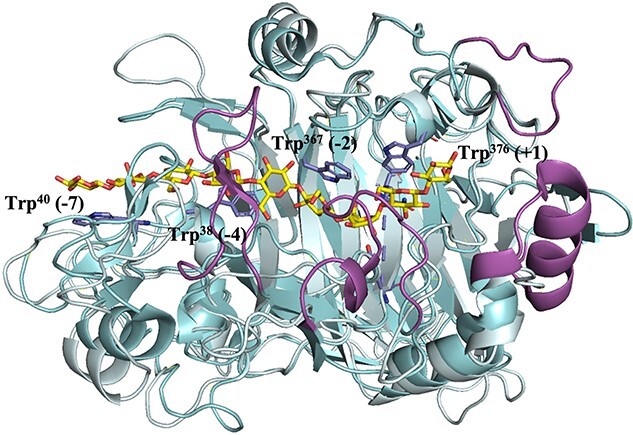
Superimposed crystal structures of the catalytic domains of cellobiohydrolase TrCel7A (cyan, PDB code 4C4C) bound to cellononaose (Knott et al., 2014) and the endoglucanase TrCel7B (pale cyan to underscore similarities, PDB code 1EG1) (Kleywegt et al., 1997). The figure shows that the catalytic domains of the enzymes are similar, but that TrCel7A has additional loops (highlighted in magenta) that create a tunnel covering the substrate-binding cleft. The substrate (yellow carbons) covers subsites +2 to −7 and interacts, in TrCel7A, with four tryptophans (side chains shown with blue carbons), Trp38 (−4 subsite) Trp40 (−7 subsite), Trp367 (−2 subsite) and Trp376 (+1 subsite). Three of these tryptophans are conserved in TrCel7B. The substrate is cleaved between the sugars bound to subsites −1 and + 1. The loops that shape the substrate-binding clefts and tunnels of GH7 cellulases have been the subject of many engineering studies. For example, Schiano-di-Cola et al. (2019) have undertaken a detailed study addressing the impact of such loops on the kinetics of substrate hydrolysis and substrate binding in TrCel7A.
