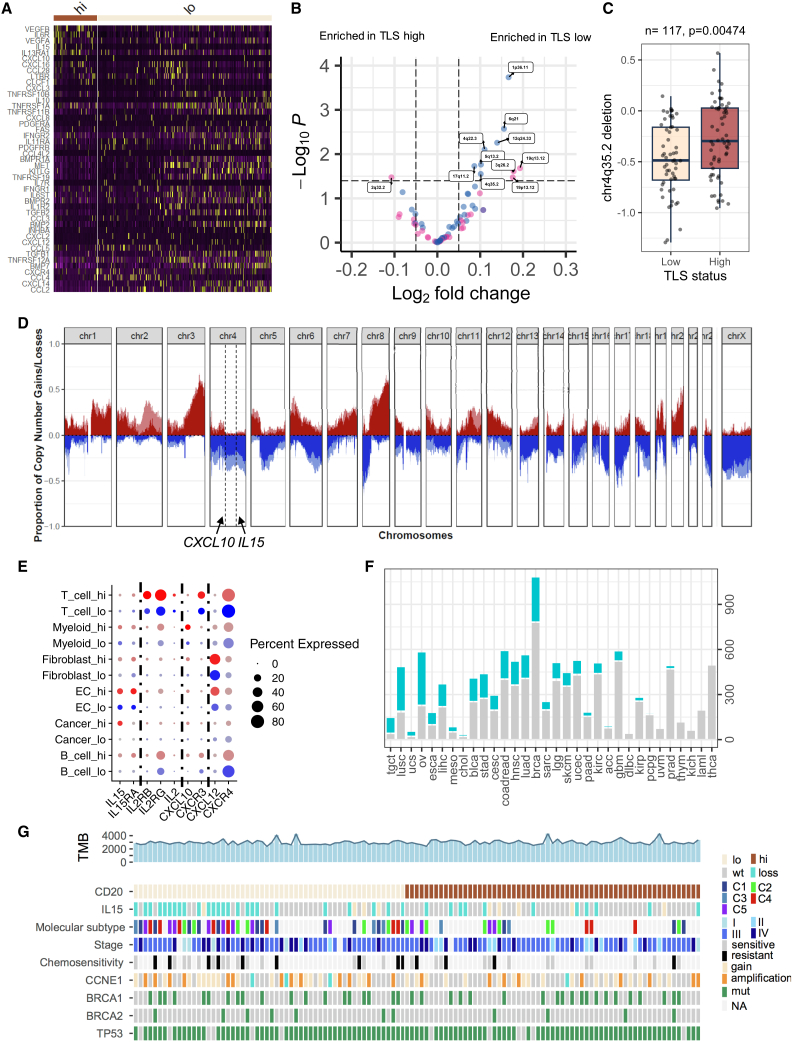
Figure 4.
Copy-number alterations dysregulate tertiary lymphoid structure formation
(A) Heatmap showing differentially expressed genes involved in cytokine signaling comparing cancer cells from TLS-high tumors with those from TLS-low tumors.
(B) Volcano plot showing genomic gains and losses associated with TLSs from the TCGA cohort. Significantly enriched genomic regions are labeled (FDR < 0.25). Horizontal line indicates FDR = 0.25. The p-values are given by a moderated t test. Pink, copy number gains; blue, copy number deletions. TLS signature was derived from the B lineage from MCPcounter.
(C) Boxplot showing association between chr4q35.2 and TLS status from the HH cohort. The p-value is given by two-tailed t test. TLS low, n = 56; TLS high, n = 61.
(D) Proportion of copy-number alterations comparing TLS-high tumors (dark red and dark blue) with TLS-low tumors (light red and light blue) in the HH cohort. The genomic locations of CXCL10 and IL15 are indicated; x axis, chromosome number; y axis, proportion of copy-number gain (red, 0 to 1) and loss (blue, 0 to −1).
(E) Dot plot showing cytokines and their receptor expression in cell subtypes. Dot color: red, cells in the TLS-high group; blue, cells in the TLS-low group. Dot size represents percentage of cells expressing the gene.
(F) Number of tumors containing IL15 deletion across cancer types; x axis, number of cases; cyan, cases with partial IL15 deletion; gray, cases without IL15 loss.
(G) Heatmap showing IL15 deletion and clinical phenotypes associated with TLS in the HH cohort. TMB, tumor mutational burden.