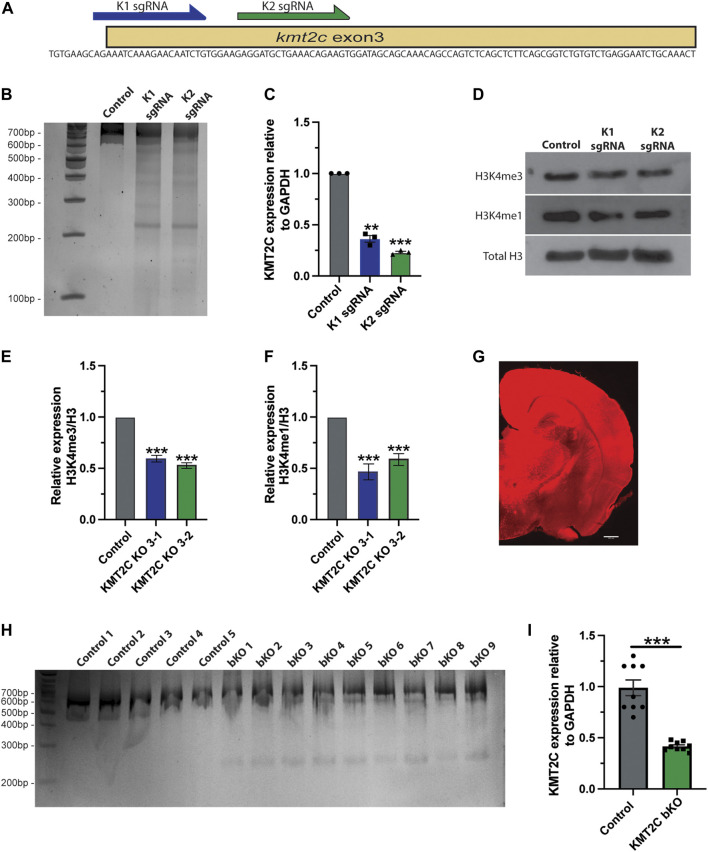
FIGURE 1.
Knockout of KMT2C by gene editing in vitro and in vivo. (A) Scheme showing the exon 3 of KMT2C genomic sequence and the positions where K1-K2 sgRNA were designed. (B) T7 endonuclease I assay from transduced cultured neurons. (C) RT-qPCR to determine expression levels of KMT2C relative to GAPDH in transduced cultured neurons. (D) Representative image of western blot analysis of total H3 histone, H3K4me3, H3K4me1. (E,F) Quantification of the relative expression of H3K4me3 (E) or H3K4me1 (F). (G) Representative image of brain slice after transduction by intracerebroventricular injection of CRISPR/Cas9. (H) T7 endonuclease I assay from transduced cortical tissue of injected animals. (I) RT-qPCR to determine expression levels of KMT2C relative to GAPDH in transduced cortical tissue of injected animals. Bars represents mean ± SEM; **p < 0.01, ***p < 0.001. Students t-test was used to determine significance compared to control condition. Scale bar = 500 μm.