FIGURE 3.
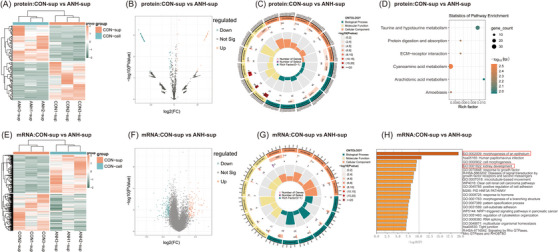
Target analysis of differentially expressed genes between the ANH and normal sEVs groups. (A) Heatmap of proteins differentially expressed between CON‐sup and ANH‐sup. One hundred sixteen differential expression proteins were identified between ANH‐sup and CON‐sup, including 56 up‐regulated and 60 downregulated proteins. (B) Volcano plots of statistical analysis of all proteins identified in CON‐sup and ANH‐sup. (C) GO analysis of proteins differentially present in CON‐sup versus ANH‐sup by circus plot. (Presenting the main 6 of biological process, cellular component and molecular function, respectively). (D) Bubble plot showing the numbers of DEG‐targeted proteins in each KEGG pathway. (E) Heatmap of mRNAs differentially expressed between CON‐sup and ANH‐sup. Eight hundred thirty‐six differential expression mRNAs were identified between ANH‐sup and CON‐sup, including 421 upregulated and 415 down‐regulated mRNAs. (F) Volcano plots of statistical analysis of all mRNAs identified in CON‐sup and ANH‐sup. (G) Circus plot showing GO analysis of mRNAs differentially present in CON‐sup versus ANH‐sup. (Presenting the main 5 of biological process, cellular component and molecular function, respectively). (H) The top 20 KEGG pathways of mRNAs targeted by the DEMs identified between CON‐sup and ANH‐sup. CON‐sup, normal fetal amniotic fluid sEVs; ANH‐sup, ANH amniotic fluid sEVs. CON‐sup, normal fetal amniotic fluid sEVs; ANH‐sup, ANH amniotic fluid sEVs.
