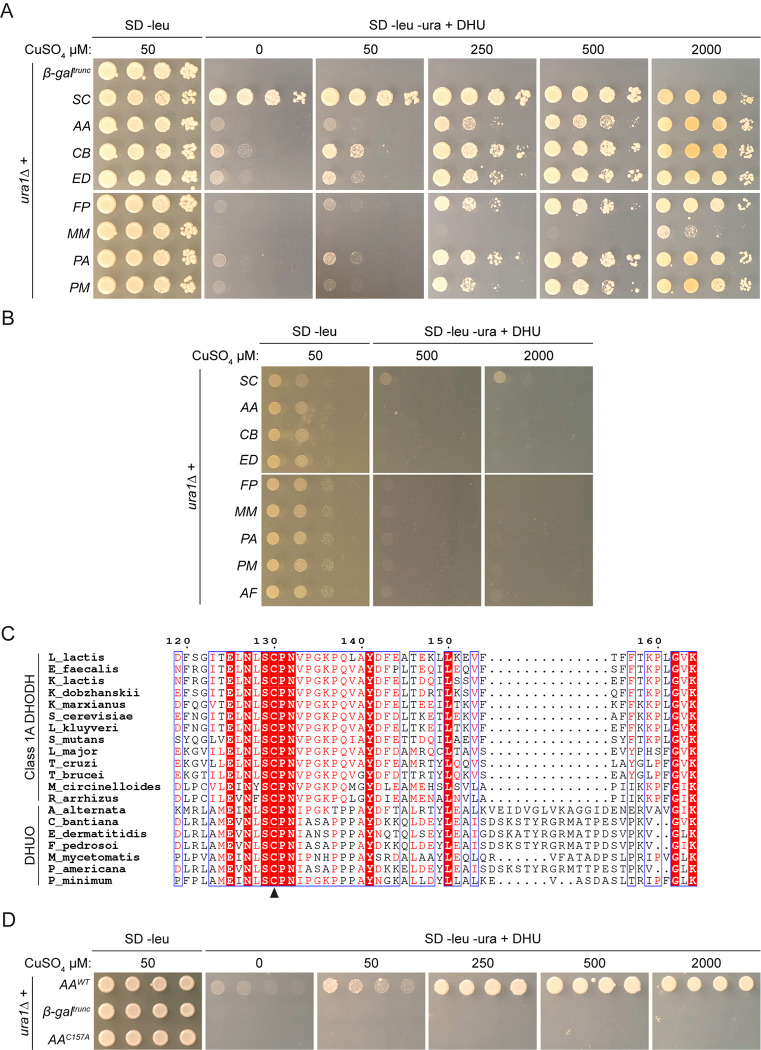
Fig 7. Dematiaceous mould putative genes have DHUO activity in the yeast model.
(A) Serial dilution assay of the negative control (β-galtrunc), confirmed class 1A DHODH from S. cerevisiae (SC) and putative genes from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells in the presence of DHU. (B) Serial dilution assay of the confirmed class 1A DHODH from S. cerevisiae (SC) and putative genes from A. alternata (AA), C. bantiana (CB), E. dermatitidis (ED), F. pedrosoi (FP), M. mycetomatis (MM), P. americana (PM) and P. minimum (PM) in ura1Δ cells under anaerobic conditions in the presence of DHU. (C) Sequence alignment of confirmed class 1A DHODH and dematiaceous DHUO. Sequences were aligned using MULTALIN [66] and displayed graphically using ESPript [86]. Alignment truncated to show only active site area. Residue numbers correspond to L. lactis, black arrowhead denotes conserved catalytic cysteine mutated. Red highlight, strict identity; red character, similarity within aligned residues; blue frame, similarity across multiple aligned residues. (D) Serial dilution assay comparing the ability of wild-type (AAWT), negative control (β-galtrunc) and cysteine mutant (AAC157A) A. alternata DHUO to complement ura1Δ cells.