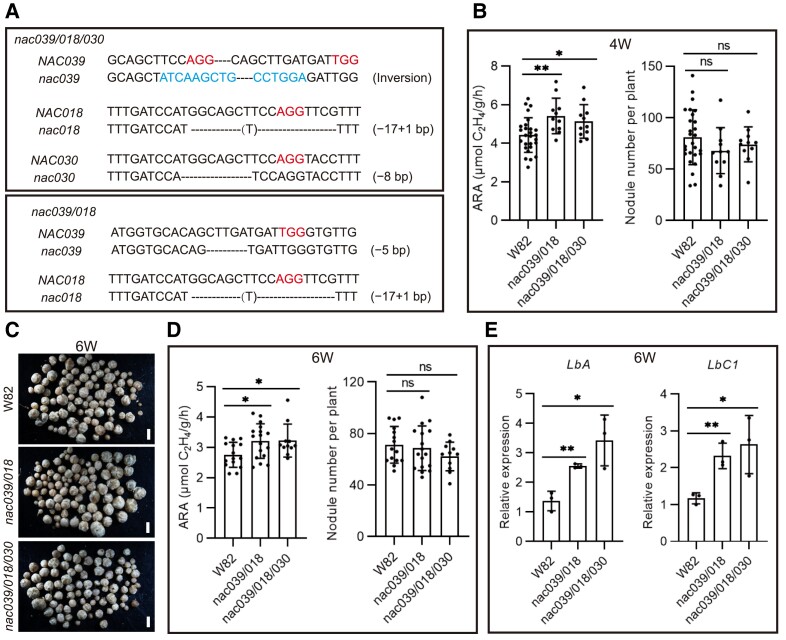
Figure 4.
Nodule number and nitrogenase activity in nac039/nac018/nac030 knockout mutant plants. A) Gene editing types in NACs-KO (knockout) stably transformed plants. An inversion between gRNA1 and gRNA3 within NAC039, a 16 bp fragment deletion at gRNA1 within NAC018, and an 8-bp deletion at gRNA1 within NAC030 were found in the nac039/nac018/nac030 triple homozygous mutant plants. A 5-bp deletion at gRNA3 within NAC039 and 16 bp deletion at gRNA1 within NAC018 were found in the nac039/nac018 double homozygous mutant plants. B) Nitrogenase activity and nodule number of NACs-KO stably transformed plants at 4 wpi. Soybean wild-type W82 was used as the control. C) Nodule phenotype of NACs-KO stably transformed plants at 6 wpi. Scale bar = 1.5 mm. D) Nitrogenase activity and nodule number of NACs-KO stably transformed plants at 6 wpi. Data shown in bar charts are means ± SD. E) Relative expression of LbA and LbC1 in hairy root nodules at 6 wpi. Three individual nodule samples were analyzed by qPCR using GmACT11 as a reference. Data shown in bar charts are means ± SD. Asterisks show significant differences determined using GraphPad Prism version 9 through unpaired Student's t-tests of comparisons with the control (*P ≤ 0.05; **P ≤ 0.01; ns, no significant difference).