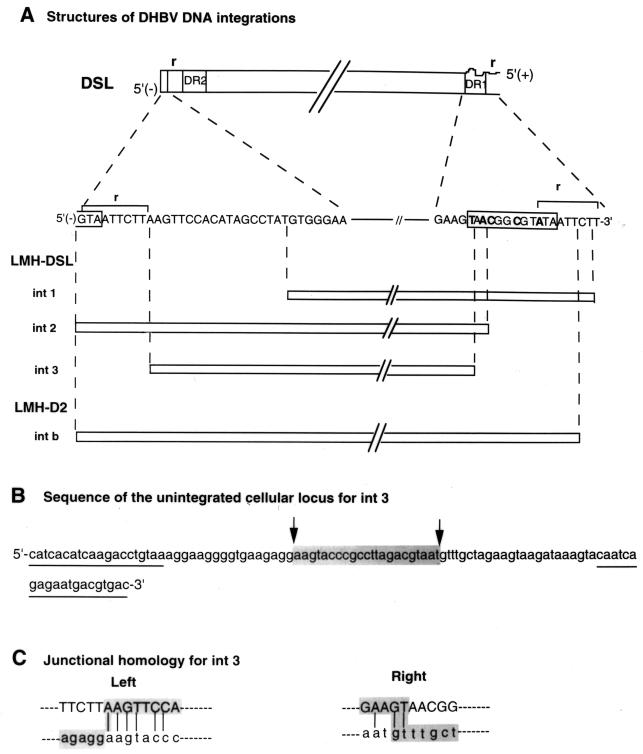
FIG. 8.
Structure of three DHBV DNA integrations from the LMH 66-1 DSL cell line. (A) Top section labeled DSL is a schematic map of a DSL DHBV virion DNA produced from LMH 66-1 DSL cell line and the specific minus-strand DNA sequences present at each end of the DSL molecule. Curvy line shows the 5′ end of the plus strand (+) of DSL DHBV DNA, which contains an 18-nucleotide RNA primer including the r-terminal redundancy region. Nucleotides in bold in DR1 indicate the mutated nucleotide changes to produce DSL DNAs. Section labeled LMH-DSL (int1, int2, and int3) is a map of three new integrations cloned from LMH-DSL subclone P1(5)-4 (integration bands seen in Fig. 6, lane 4). Vertical dashed lines denote left and right viral junctions with cellular DNA. LMH-D2 intb is a previously reported WT DHBV integration (9) with a structure similar to that of DSL DNA. Complete DHBV integrations and their immediate cellular flanking DNA were sequenced manually or by an automatic sequencer using oligonucleotides derived from the DHBV genome and cellular DNA. Slashes (//) in the diagram represent the uninterrupted DHBV genome. (B) The unintegrated cellular locus for LMH 66-1 DSL int3 was isolated by PCR from the untransfected LMH cell line using oligonucleotides derived each from the left and right flanking cellular DNA of int3. One specific fragment was amplified by PCR from the LMH cell line from the oligonucleotide primers (underlined) and was sequenced. The shaded sequence represents region of the unintegrated locus DNA that was not present in the int3 flanking cellular DNA. The two arrows indicate possible sites of deletion of cellular DNA during int3 integration. (C) DHBV DNA at the junction site of int3 (upper-case letters) and cellular genomic DNA sequences across the left and right junctions of int3 (lower-case letters) were aligned for comparison. Vertical lines denote the homology between the viral and cellular DNA sequences at the junctions. The shaded sequences are int3 junctional sequences.