Figure 6. RIF1–SHLD3 interaction is essential for shieldin function.
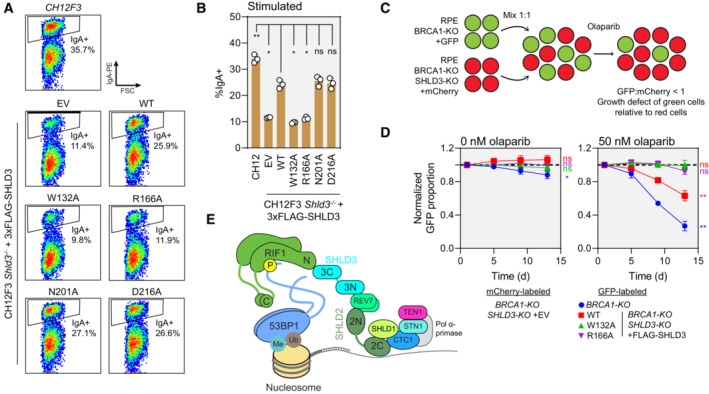
- Representative flow cytometry density plots (from three biologically independent experiments) measuring IgA expression in stimulated CH12F3 Shld3 −/− cells stably expressing FLAG‐tagged SHLD3 alanine substitution variants through lentiviral transduction. Values shown are %IgA+ cells. WT—wild‐type. EV—empty vector. FSC—forward scatter. See also Fig EV5C–E.
- Quantification of class switch recombination in stimulated CH12F3 Shld3 −/− cells stably expressing the indicated FLAG‐SHLD3 variants through lentiviral transduction. Bars represent mean ± s.d., n = 3 biologically independent experiments. Analysis was performed using one‐way ANOVA followed by the Dunnett's multiple comparisons test against Shld3 −/− cells complemented with wild‐type SHLD3. **P < 0.01, *P < 0.05, ns P > 0.05.
- Schematic of the two‐color competitive growth assay. The indicated RPE cell lines transduced with lentivirus encoding either eGFP or mCherry. eGFP‐ and mCherry‐labeled cells were mixed 1:1 and treated with olaparib, and the relative number of labeled cells was monitored over time by automated fluorescence microscopy. eGFP:mCherry ratios are normalized to the value 1‐day postplating. An eGFP:mCherry ratio < 1 indicates slower growth of the eGFP‐expressing cell line relative to the mCherry‐expressing cells.
- Competitive growth assay of RPE BRCA1‐KO SHLD3‐KO cells complemented with 3xFLAG‐SHLD3 variants. Growth of the indicated eGFP‐expressing cells was compared with mCherry‐expressing RPE BRCA1‐KO SHLD3‐KO cells transduced with empty vector after mixing 1:1 in 24‐well plates. Plates were imaged 1, 5, 9, and 13 days after plating. Dotted black lines represent an eGFP:mCherry ratio of 1. Points represent mean ± s.d. (n = 3 biologically independent experiments). Analysis was performed using a one‐sample t‐test against a hypothetical eGFP:mCherry ratio value of 1 for the day 13 time point. **P < 0.01, ns P > 0.05. See also Fig EV5F.
- Schematic of the molecular basis for 53BP1‐RIF1‐shieldin‐CST recruitment to sites of DNA damage.
Source data are available online for this figure.
