Figure 4. AS L1 RNAs facilitate meshwork‐like organization of MATR3 proteins.
-
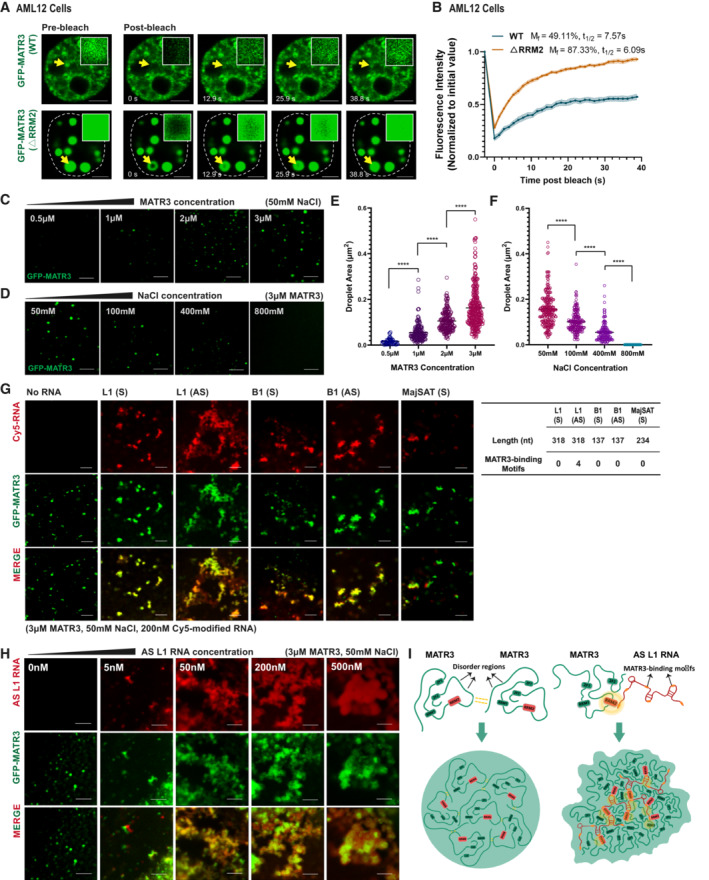
ARepresentative images of the GFP‐MATR3 (WT/▵RRM2) FRAP experiments in AML2 cells. Solid arrows indicate the bleached points. Scale bars, 5 μm.
-
BThe fluorescence recovery curve of the GFP‐MATR3 (WT/▵RRM2) corrected for fluorescence decay. Data are expressed as the mean ± s.e.m. (WT, n = 11; ▵RRM2, n = 11).
-
CRepresentative images of droplet formation assays with different concentrations of GFP‐MATR3 proteins. NaCl concentration, 50 mM. Scale bars, 5 μm.
-
DRepresentative images of droplet formation assays with different NaCl concentrations. GFP‐MATR3 protein concentration, 3 μM. Scale bars, 5 μm.
-
EAreas of MATR3 protein droplets formed in different protein concentration (3 μM: n = 315; 2 μM: n = 196; 1 μM: n = 165; 0.5 μM: n = 50). The P‐values were calculated using unpaired two‐tailed Student's t‐test; ****P < 0.0001.
-
FAreas of MATR3 protein droplets formed in different NaCl concentration (50 mM: n = 248; 100 mM: n = 196; 400 mM: n = 127; 800 mM: none). The P‐values were calculated using unpaired two‐tailed Student's t‐test; ****P < 0.0001.
-
G(Left) Representative images of droplet formation assays by GFP‐MATR3 with different in vitro‐transcribed RNAs (Sense L1, Antisense L1, Sense B1, Antisense B1, MajSAT). RNA concentration, 200 nM. GFP‐MATR3 protein concentration, 3 μM. NaCl concentration, 50 mM. (Right) The table showing the nucleotide number and the number of MATR3‐binding motifs on the in vitro‐transcribed RNAs. Scale bars, 5 μm.
-
HRepresentative images of droplet formation assays by GFP‐MATR3 with different concentration (0, 5, 50, 200, 500 nM) of AS L1 RNAs. GFP‐MATR3 protein concentration, 3 μM. NaCl concentration, 50 mM. Scale bars, 5 μm.
-
ISchematic representation for MATR3‐MATR3 droplets formation and MATR3‐AS L1 RNA meshwork formation.
Source data are available online for this figure.
