Abstract
Background
The lack of murine glioblastoma models that mimic the immunobiology of human disease has impeded basic and translational immunology research. We, therefore, developed murine glioblastoma stem cell lines derived from Nestin-CreERT2QkL/L; Trp53L/L; PtenL/L (QPP) mice driven by clinically relevant genetic mutations common in human glioblastoma. This study aims to determine the immune sensitivities of these QPP lines in immunocompetent hosts and their underlying mechanisms.
Methods
The differential responsiveness of QPP lines was assessed in the brain and flank in untreated, anti-PD-1, or anti-CTLA-4 treated mice. The impact of genomic landscape on the responsiveness of each tumor was measured through whole exome sequencing. The immune microenvironments of sensitive (QPP7) versus resistant (QPP8) lines were compared in the brain using flow cytometry. Drivers of flank sensitivity versus brain resistance were also measured for QPP8.
Results
QPP lines are syngeneic to C57BL/6J mice and demonstrate varied sensitivities to T cell immune checkpoint blockade ranging from curative responses to complete resistance. Infiltrating tumor immune analysis of QPP8 reveals improved T cell fitness and augmented effector-to-suppressor ratios when implanted subcutaneously (sensitive), which are absent on implantation in the brain (resistant). Upregulation of PD-L1 across the myeloid stroma acts to establish this state of immune privilege in the brain. In contrast, QPP7 responds to checkpoint immunotherapy even in the brain likely resulting from its elevated neoantigen burden.
Conclusions
These syngeneic QPP models of glioblastoma demonstrate clinically relevant profiles of immunotherapeutic sensitivity and potential utility for both mechanistic discovery and evaluation of immune therapies.
Keywords: Anti, CTLA, 4, anti, PD, 1, glioblastoma, immunotherapy
Key Points.
Nestin-CreER T2 Qk L/L ; Trp53 L/L ; Pten L/L glioblastoma lines replicate the immunotherapy resistance of clinical disease.
Programmed death ligand 1upregulation mediates checkpoint resistance in the brain but not the flank.
High neoantigen density sensitizes immune microenvironments of sensitivity to immunotherapy in both brain and flank potential.
Importance of the Study.
To understand the mechanisms engaged by glioblastoma in vivo to suppress T cell immunity and immunotherapy, as well as to develop strategies to overcome these obstacles translationally, pre-clinical models of glioblastoma which recapitulate its key clinical immune features are critically needed. We present syngeneic mouse models of glioblastoma derived from Nestin-CreERT2QkL/L; Trp53L/L; PtenL/L (QPP) mice with a spectrum of checkpoint blockade sensitivity ranging from complete resistance to sensitivity. We show how the differences in sensitivity to checkpoint blockade between these models or between the same model growing on the flank versus in the brain can be used to identify elements of the tumor microenvironment central to the establishment of glioblastoma immunotherapy resistance. These models provide a genomically and immunologically relevant pre-clinical platform for discovery and screening of new immune-based therapies for glioblastoma.
Glioblastoma survival remains poor at an average of only 15 months. Despite the success of T cell immune checkpoint blockade (ICB) therapy in cancers such as melanoma, clinical trials have established that blockade of programmed death 1 (PD-1) fails to improve survival in glioblastoma.1 In the broadest sense, immunotherapy resistance in glioblastoma may originate at the level of the glioma-stem cells and can be associated with both recruitment of a suppressive tumor microenvironment (TME) and the inherent genomic heterogeneity, of this cancer.2,3 Of note, heterogeneity in GBM originates more from epigenomic variation than from an elevated density of DNA mutation which could foster enhanced immunogenicity. Myeloid-derived suppressor cells, tumor-associated macrophages, and microglia act in concert in glioblastoma to cripple T-cell immunity.4 Beyond the myeloid stroma, inhibitory checkpoint molecules such as programmed cell death ligand 1 (PD-L1) can be highly enriched across the glioblastoma TME and negatively impact both local immune activation and overall prognosis in patients.5
Despite the clinical failure of immunotherapy for glioblastoma, the widely utilized murine model GL261 is highly immune sensitive and readily cured by blockade of PD-16 (Supplementary Figure S1). This incongruity between encouraging pre-clinical findings in GL261 and the lack of responses in patients indicates that new and more robust models are needed that more accurately reflect the immune composition and therapeutic (in)sensitivity of human disease. On the contrary, mice engineered to spontaneously develop glioblastoma in situ using the RCAS/Ntv-a system require little to no genetic “second hits” beyond the driver oncogenes (eg, PDGF-β, BCL-2) and therefore, likely lack immune targetable mutations which are present, albeit at a modest frequency in human disease.7 A recent model, SB28, showed resistance to the PD-1 and CTLA-4 blockade was generated de novo using transposon-mediated transfection to overexpress oncogenes (eg, NRas, PDGF) and downregulate tumor suppressor gene TP53. Despite its resistance, the mutation heterogeneity of SB28, similar to the engineered mouse model driven by specific oncogenes was also found more concentrated in certain pathways.8,9
Glioblastomas frequently suppress or delete the tumor suppressor gene Quaking (QKI), and this depressed expression highly correlates with poor patient survival.10 Analysis of TCGA datasets revealed 20% deletion of QKI in 420 samples and methylation analysis revealed 20% methylation of QKI in 250 profiles.11 Multiple additional studies have found QKI deletion in ~30% of GBM.12,13QKI deletion also yields dysregulated mitophagy and induces reactive oxygen species and DNA damage, all of which act to foster genomic instability and potential secondary genetic mutations. We have previously reported that the brain tumors which arise in mice deleted for QKI in addition to PTEN and TP53 (Nestin-CreERT2QkL/L; Trp53L/L; PtenL/L or QPP) demonstrate histopathological heterogeneity and a transcriptomic profile resembling of all 4 subtypes of human glioblastoma.10
In this study, we establish 4 murine syngeneic QPP tumor lines derived from these mice and determine their distinct immune sensitivities to T-cell checkpoint inhibitors in both the brain and subcutaneous niches. Emblematic of the potential utility of these novel glioblastoma pre-clinical models, we utilize comparative analysis of the immune microenvironments and mutational profiles of T cell ICB sensitive versus resistant clones to understand the factors that drive immune privilege in the CNS. These novel glioblastoma mouse models which replicate both the histopathological and transcriptomic features and immune checkpoint insensitivity of human disease may provide critical tools for both understanding the immunobiology of glioblastoma, as well as for vetting potential immunotherapeutic approaches to select the most promising for clinical advancement.
Materials and Methods
Animals
C57BL/6J mice and RagKO (B6.129S7-Rag1tm1Mom/J) mice were from The Jackson Laboratory (Bar Harbor, ME) and housed according to AAALAC and NIH standards. All experiments were conducted under protocols approved by the MD Anderson Cancer Center (MDACC) Institutional Animal Care and Use Committee.
Cell Lines and Reagents
QPP stem cells were derived from QPP mice10 and cultured in DMEM/F12 media with B-27 supplement (Gibco), epidermal growth factor, and fibroblast growth factor (STEMCELL technologies).10 Among the QPP cell lines tested in this study, only QPP7 expresses GFP due to its genetic background; however, GFP is known to have minimal immunogenicity in C57BL6 mice.14 Arginine-deficient media was used for arginine auxotrophy experiments. Interferon-γ (PeproTech, 200 ng/mL) was used to induce PD-L1 expression in QPP in vitro. GL261 was obtained from Dr Amy Heimberger (MDACC) and cultured in DMEM with 15% fetal bovine serum and non-essential amino acids (Gibco).
Flank Tumor Growth
A total of 1 × 106 QPP cells were implanted in 30% Matrigel (Corning) on the flank of male C57BL/6J or RagKO mice. Tumor dimensions were measured with calipers.
QPP Line Phenotyping
QPP cell lines were stained for surface markers CD171 (Miltenyi Biotec), α2β5, CD44, CD133 (Biolegend), Sca-1 (Thermo), and PD-L1 and PD-L2 (BD Biosciences). Tumors extracted from tumor-challenged mice were digested with Collagenase H (Sigma) and DNase (Roche) into single-cell suspensions as described.15 Cells were stained for the above surface markers in addition to CD45.2 (Biolegend) to exclude immune cells.
Immunotherapy Studies
For s.c. tumors, 1 × 106 QPP4, 5, 7, or 8 were implanted on the left flank of male C57BL/6J mice. For intracranial studies, 1 × 105 cells (QPP7 and GL261), 2 × 105 cells (QPP4 and 5), and 3 × 105 cells (QPP8) were mixed with 50% methylcellulose (Acros Organics) to a total volume of 5 μl—optimal cell doses for each line were determined as the minimum necessary for complete engraftment. Cells were injected into the right striatum of C57BL/6J mice, at approximately 2 mm anterior and 2 mm right lateral to the Bregma at a depth of 4 mm, using a stereotactic device. Therapeutic CTLA-4 (9H10, 100 μg/dose) and PD-1 (29F.1A12, 200 μg/dose or RMP1-14, and 250 μg/dose) antibodies were purchased from BioXcell or Leinco. PD-1 antibodies were sourced based on availability and are functionally equivalent at these doses. Mice received 3 doses of antibody or PBS i.p. on days 7, 10, and 13. Flank tumor volume was measured with calipers to 1000 mm3 and brain tumor mice were monitored for morbidity and/or survival.
Flow Cytometry Analysis of Tumor Infiltrate
For flank tumors, 1 × 106 QPP8 cells were implanted followed by 3 doses of antibody given i.p. every 3 days. For brain tumors, 1 × 105 (QPP7, GL261) or 5 × 105 (QPP8) cells were inoculated in 5μl of 30% Matrigel into the right striatum of C57BL/6J mice. Three antibody treatments or PBS were given i.p. every 3 days starting on day 5 for GL261, on day 33 for QPP7, and on day 48 for QPP8. All mice were sacrificed 2 days after the last treatment. Tumors were digested with Collagenase H and DNase and their cell numbers were counted. After the removal of myelin via Percoll gradient (Sigma) for brain tumors, live immune cells were enriched by Ficoll gradient centrifugation (Sigma; 1119 density). We stained the cells using our designed panel for immune phenotypic markers. Expression data were collected on a 20-parameter BD LSRII flow cytometer and analyzed with FlowJo Version 10. Immune subsets, staining panel, and method are detailed in Supplementary Figure S2.
Analysis of Genomic Alterations in QPP Cell Lines
Genomic DNA was extracted from QPP lines using the GeneJET Genomic DNA Purification Kit (Thermo). Novogene performed whole exome sequencing of the samples using the paired-end 150 methods with a depth of 50–100X and coverage of > 99%.
Prediction of Neoantigens in QPP7 and QPP8
Somatic single-nucleotide mutations were identified using MuTect.16 To determine wild type and mutant peptide binding affinities to class-I MHC alleles (H-2-Db and H-2-Kb) we used the NetMHC program,17 which applies artificial neural networks to predict peptide-MHC binding. A peptide was considered to be a binder to MHC if the predicted binding affinity was <1000 nM.
Determination of TCR Repertoire in Intratumoral T cells
A total of 1 × 105 (QPP7) or 5 × 105 (QPP8) cells were inoculated in 5μL of 30% Matrigel into the right striatum of C57BL/6J mice. Tumors were collected 33 and 48 days after implantation for QPP7 and 8, respectively. The immune cells were isolated using the method described above and stained with CD45, CD3, CD4, and CD8 fluorescently labeled antibodies. CD4 and CD8 T cells were enriched using fluorescence-activated cell sorting at MDACC The flow cytometry and cellular imaging core facility. DNA were purified using DNeasy Blood and Tissue Kit (Qiagen) and TCR sequencing was performed by ImmunoSeq.
Statistical Analysis
All statistics were calculated using GraphPad Prism. Survival was generated in Kaplan–Meier plots and survival difference across groups was determined by Log-Rank (Mantel-Cox) test. For tumor infiltrate analysis, a 2-sided student’s t-test applying Welch’s correction for unequal variance was used to determine statistical significance. P-values < .05 were considered significant.
Heat Maps
All heat maps show the log of fold changes of the indicated ratio or specific parameter MFI compared to the QPP line and tissue site-matched control group. Each ratio or MFI fold change in the treatment groups was normalized to the mean of the same parameter of the matching PBS control group. The normalized results in each independent experiment were then combined. For each parameter, student’s t-test was used to compare the combined normalized fold changes in the treatment groups with the combined fold changes in the PBS group (mean = 1). “*” indicate differences with a P-value < .05.
Results
Four QPP Tumor Lines Grow Progressively in C57BL/6 Mice
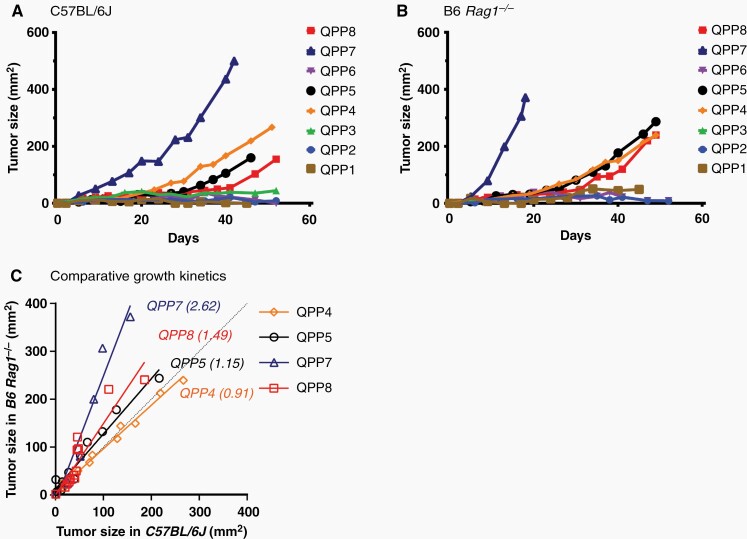
We tested 8 glioma-stem cell lines derived from Nestin-CreERT2QkL/L; Trp53L/L; PtenL/L (QPP) mice for growth potential following s.c. implantation on the flank of immunocompetent C57BL/6J and immune deficient B6 Rag1−/− (RagKO) mice. Four of these QPP cell lines (QPP4, 5, 7, and 8) grew progressively in both settings suggesting their potential utility as transplantable models of glioblastoma (Figure 1A and B). In this context, QPP7 grew most rapidly among the lines. QPP7 also showed the greatest attenuation of growth rate in wild-type versus RagKO mice, however, suggesting that it may have the most sensitivity to adaptive immunity (Figure 1C).
Figure 1.
Four QPP lines grow in both immune-deficient and immunocompetent mice. 1 × 106 cells of each QPP cell line were subcutaneously implanted in (A) C57BL/6J (n = 4) and (B) B6 Rag1 knockout (RagKO) mice (n = 2) and their growth was measured with calipers. Each curve and error bar represents the mean and SEM of each QPP line. A comparison of growth kinetics between C57BL/6J and B6 Rag1 knockout is also demonstrated (C).
QPP Tumor Lines Retain a Subset of Glioma-stem Cell Markers but Express Few Immune Checkpoint Ligands
All QPP lines can form neurospheres in serum-free media—a cardinal feature of glioma-stem cells. Using flow cytometry analysis of common glioma-stem cell markers, we found that all 4 selected QPP lines prominently express classical stem cells markers such as CD44, CD171, and A2B5 with some variation (ie, low A2B5 but some Sca-1 in QPP7 and lower CD171 in QPP8), which are critical in tumor initiation, maintenance, and invasion (Supplementary Figure S3).18–20 While expression of immune inhibitory checkpoint ligands such as PD-L1 by glioblastoma has been described,21 the extent of checkpoint ligand expression by tumor-initiating glioma-stem cells remains poorly described. We found little to no baseline expression of T cell checkpoint ligands, such as PD-L1 or PD-L2, on the syngeneic QPP cell lines in vitro (Supplementary Figure S3A, B). Upon stimulation with interferon (IFN)-γ, however, all of the QPP lines upregulate moderate to high levels of PD-L1. QPP7 also had the highest level of induced PD-L1 expression suggesting potential sensitivity to PD(L)-1 blockade (Supplementary Figure S4).
QPP Gliomas are Differentially Sensitive to Blockade of the T Cell Checkpoints CTLA-4 and PD-1
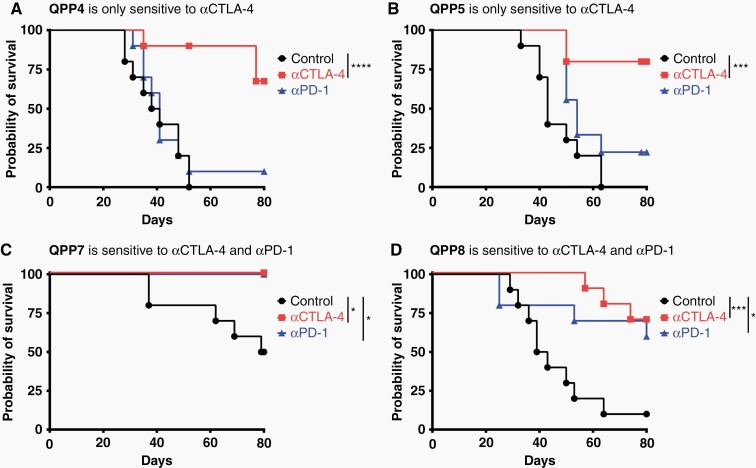
The capacity to grow both in the brain and on the flank can be advantageous both for investigating comparative biology and for therapeutic screening applications. To assess the immunotherapeutic sensitivity of the 4 QPP lines, we implanted these lines in the flank and treated them systemically with either anti-PD-1 or anti-cytotoxic T lymphocyte-associated molecule 4 (CTLA-4) on days 7, 10, and 13. As predicted from its PD-L1 expression and slower growth in wild-type versus RagKO mice, the QPP7 tumor line proved to be highly sensitive to PD-1 blockade (Figure 2 and Supplementary Figure S5). In contrast to QPP7 and the prevalent glioma model GL261, however, the QPP4 and QPP5 lines were resistant to anti-PD-1 therapy. All 4 lines did retain significant sensitivity to CTLA-4 blockade. QPP8 shows an interesting profile of response to anti-CTLA-4, and sometimes anti-PD-1, in which nearly complete initial tumor regression gives way to subsequent progression. Few transplantable mouse models exist which replicate this pattern of response, remission, and relapse.22
Figure 2.
T cell checkpoint blockade sensitivity of flank-implanted QPP cell lines. 1 × 106 cells were subcutaneously implanted on day 0 and mice were treated i.p.with anti-PD-1 (29F.1A12 or RMP1-14) or anti-CTLA-4 (9H10) antibody or PBS on days 7, 10, and 13. Tumors were measured with calipers, and a tumor measuring more than 800 mm3 is considered a death event. Each curve represents the survival curve from 2 experiments of 5 mice per group.
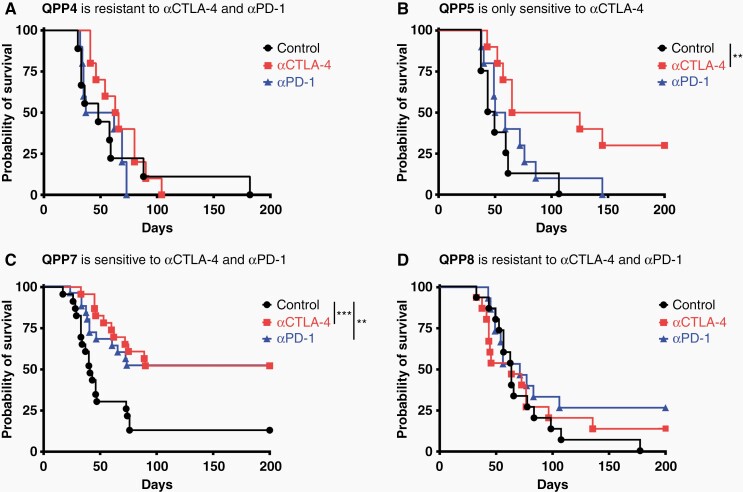
We evaluated the essential property of these QPP gliomas by orthotopically implanting each line and treating them with i.p. anti-PD-1 or anti-CTLA-4 on days 7, 10, and 13. All 4 lines progressed to fatal glioblastoma without treatment and manifested diverse sensitivities to systemic ICB (Figure 3). QPP7 grew most aggressively (median survival 41.0 days) and QPP8 most slowly (median survival 63.0 days). Like human glioblastoma, QPP4 and QPP8 tumors are resistant to both ICB. QPP5 and QPP7 both responded to CTLA-4 blockade (P < .01 and P < .001, respectively); however, only QPP7 also trended toward anti-PD-1 sensitivity (P = .01) (Figure 3). In this model, the unique combination of Pten, p53, and Qki loss appears capable of imprinting varied patterns of immune sensitivities likely reflecting underlying genomic heterogeneity.
Figure 3.
QPP lines engraft orthotopically and are differentially sensitive to checkpoint blockade. 1 × 105 (QPP7), 2 × 105 (QPP4 and 5) and 3 × 105 (QPP8) cells were stereotactically implanted into the right striatum of C57BL/6J mice. On days 7, 10, and 13, mice were treated i.p. with anti-PD-1 (29F.1A12 or RMP1-14) or anti-CTLA-4 (9H10) antibodies or PBS. Survival and significance are shown based on the Log-Rank (Mantel-Cox) test. Two (QPP4 and 5) to 3 (QPP7 and 8) experiments were performed with 4–5 mice per group.
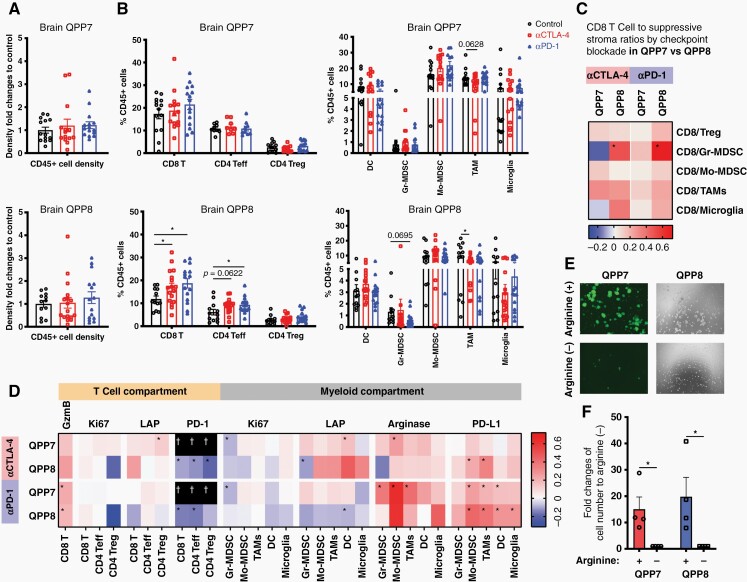
Elevated ratios of CD8 T cells relative to suppressive T and myeloid cells support CD8 response in the flank versus resistance in the brain
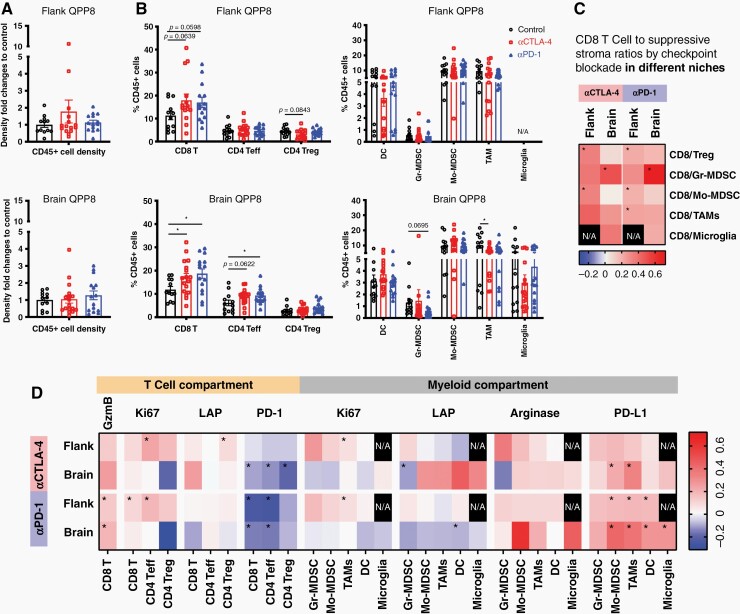
We investigated changes in the QPP8 immune infiltrate when treated in the flank (sensitive) versus in the brain (resistant). Surprisingly, we found similar infiltration of CD45+ immune cells in response to either CTLA-4 or PD-1 blockade regardless of implantation site suggesting conserved primary mechanisms of immune suppression (Figure 4A). Although brain QPP8 fails to respond therapeutically to ICB, CD8 T cell frequencies in the tumor are more significantly increased in comparison to when QPP8 is implanted subcutaneously. In addition, levels of tumor-associated macrophages decline in response to ICB in the brain but not the flank; however, any therapeutic benefit here may be offset by the substantial microglial composition in the brain that remains unchanged by ICB (Figure 4B). This CTLA-4 antibody can deplete Tregs in peripheral tumors, which we observed as a trend (P = .084) that may contribute to the response in the flank (Figure 4B). Consistent with this effect, we observed significantly enhanced ratios of CD8 T cells relative to Treg in QPP8 in the flank with both anti-CTLA-4 and anti-PD-1 therapy that, as the sum of both greater CD8 T cell infiltration and greater Treg depletion, create a more pro-inflammatory ICB response than is mobilized in the brain (Figure 4C and Supplementary Figure S6A). Furthermore, ratios of CD8 T cells relative to suppressive myeloid stroma all appear elevated following ICB in flank QPP8 (P < .05 for CD8/Mo-MDSC and/tumor-associated macrophages for αPD-1 and CD8/Mo-MDSC for αCTLA-4) indicative of pro-inflammatory conditioning of the TME. In contrast, these enhanced ratios were not observed in brain QPP8 except for an improved ratio of CD8 T cells to granulocytic MDSC (Gr-MDSC). Of note, Gr-MDSC are the least abundant myeloid population in QPP8 regardless of the site of implantation. The capacity of ICB to reshape the cellular composition of the flank, but not brain, QPP8 tumors to favor antitumor immunity partially explains QPP8 immunotherapy resistance in the brain. Next, we explored how changes in the functional phenotypes of these populations contribute sensitivity to immunotherapy.
Figure 4.
Comparative infiltrates analysis of flank versus brain implanted QPP8. 1 × 106 cells and 5 × 105 cells of QPP8 were implanted in the flank or brain respectively. Mice received 3 doses of i.p. anti-CTLA-4 (9H10) or anti-PD-1 (RMP1-14) antibody or PBS every 3 days and were sacrificed 2 days after the last treatment. Tumor-infiltrating immune cells were extracted, enriched, and stained with phenotypic and functional markers before analysis by flow cytometry. Immune infiltrate density fold changes compared to the control group (A) and T cell subset frequencies (B) in response to T cell checkpoint blockade in both flank and brain tumors are shown as mean ± SEM. Fold changes of CD8 T cells over suppressive subset ratios (C) and functional marker MFI fold changes compared to the matched control group for each immune subset in the indicated site were calculated and presented on a log scale (D). Data reflect 3 experiments with 3–8 mice per group. (*P < .05 by Student’s t-test).
In flank QPP8, increased Granzyme B and Ki67, and decreased PD-1 expression on CD8 T cells in response to PD-1 blockade (P < .05) together suggest improved cytotoxicity and fitness (Figure 4D and Supplementary Figure S6B). Effector CD4 T cells also show elevated expansion (ie, Ki67) following blockade of either checkpoint. A subset of these improvements to T cell fitness are also found in brain QPP8, where PD-1 blockade results in enhanced Granzyme B expression and both blockades increase PD-1 downregulation. A critical difference between the flank and brain, however, occurs at the level of T cell proliferation. Whereas T cells expand in response to checkpoint blockade in subcutaneous QPP8, in the brain their proliferation remains static or even declines. This suggests either the presence of additional mechanisms of dominant T cell proliferative suppression in the brain, and/or that checkpoint antibodies no longer protect the T cells once they reach the QPP8 TME. Consistent with these findings, the highly ICB-sensitive GL261 glioma model shows an immune infiltrate pattern more similar to flank QPP8 than to resistant brain QPP8 (Supplementary Figure S7). In brain GL261, like flank QPP8, we find, as others have reported, that Granzyme B and Ki67 significantly increase following PD-1 blockade in the CD8 and CD4 effector compartments, while PD-1 expression in the CD4 effector compartment decreases.6
Across the myeloid stroma of both flank and brain QPP8, ICB triggers the upregulation of PD-L1 (Figure 4D and Supplementary Figure S6C). The most significant induction occurs in brain QPP8 for both CTLA-4 and PD-1 blockade perhaps contributing to the immunotherapy resistance observed in this site. As our analysis revealed signs of increased T cell effector function (ie, elevated Granzyme B) with ICB, adaptive PD-L1 upregulation may be triggered, at least in part, by increased T cell effector cytokine production (eg, IFN-γ). Changes in TGF-β (LAP, latency-associated peptide) and arginase between flank and brain QPP8 were either inconsistent between CTLA-4 and PD-1 blockade (LAP) or did not reach statistical significance (arginase). We cannot rule out that the enhanced immune resistance of brain QPP8 results, at least in part, from the expression of arginase by microglia in brain QPP8 while absent in the flank (Figure 4D). Despite the significant induction of PD-L1 in flank QPP8, ratios of CD8 T cells to nearly all suppressive myeloid stromal populations increase in the presence of PD-1 blockade and, to a lesser extent, CTLA-4 blockade (Figure 4C). In contrast, brain QPP8 CD8 to suppressor ratios are mostly unaffected by ICB. These findings suggest that PD-1 blockade can successfully counteract the PD-L1 upregulation observed in flank QPP8, but that the antibody may not effectively block PD-L1 induced by brain QPP8, perhaps due to limited bioavailability within the brain TME.23
QPP7 Remains Checkpoint Sensitive in the CNS Despite Adaptive Upregulation of Arginase
Comparative analysis of the TME of resistant brain QPP8 versus sensitive flank QPP8 indicated the importance of lack of ICB-induced upregulation of T cell proliferation and adaptive upregulation of PD-L1 on the myeloid stroma as potentially key factors driving immunotherapy resistance in glioblastoma. While the QPP lines share driver mutations, the loss of Qki drives histopathological heterogeneity that yielded lines with substantially divergent immune sensitivity suggesting underlying genomic heterogeneity. To understand the influence of these alterations on the glioblastoma TME, we performed comparative analysis of checkpoint-sensitive QPP7 and resistant QPP8 in the brain. We did not see significant changes in immune infiltration following CTLA-4 and PD-1 blockade in brain QPP7 (Figure 5A and B). In contrast, effector T cells expanded and tumor-associated macrophages decreased in response to ICB in poorly responsive QPP8 (Figure 5B). Regardless of therapy, QPP7 contained approximately three-fold higher frequencies of antigen-presenting dendritic cells versus QPP8 which might suggest a critical lack of antigen presentation or local T cell support that supersedes any ICB benefits to the T cell compartment. Although anti-PD-1 sensitive, brain QPP7 ratios of CD8 versus suppressive T and myeloid cells improved less in response to PD-1 blockade than those in resistant QPP8 (Figure 5C). Similarly, CTLA-4 blockade generated a more pro-inflammatory profile in brain QPP8 than in sensitive QPP7 where ratios of CD8 T cells versus Gr-MDSC and microglia actually declined relative to untreated controls. As immune sensitivity of QPP7 could not be explained by these observations, we examined phenotypic changes to key T cell and myeloid populations in response to therapy.
Figure 5.
QPP7 remains checkpoint sensitive in the CNS despite adaptive upregulation of Arginase. 1 × 105 QPP7 cells or 5 × 105 QPP8 cells were implanted in the right striatum. Mice received 3 doses of i.p. anti-CTLA-4 (9H10), anti-PD-1 (RMP1-14) antibody, or PBS every 3 days and sacrificed 2 days after the last treatment. Tumor-infiltrating immune cells were extracted, enriched, and stained for phenotypic and functional markers before analysis by flow cytometry. Immune infiltrate density fold changes compared to the control group (A) and T cell subset frequencies (B) in response to T cell checkpoint blockade in both QPP7 and QPP8 brain tumors are shown as mean ± SEM. Fold changes of CD8 T cells over suppressive subset ratios (C) and functional marker MFI fold changes compared to the matched control group for each immune subset in the indicated site were calculated and presented on a log scale (D). Data reflect 3 experiments with 3–8 mice per group (*P < .05 by Student’s t-test). 6 × 105 QPP7 and QPP8 cells (dash line) were seeded in 10 mL of media either with or without arginine and were incubated for 6 days before image analysis by fluorescent and light microscopy (E) and viable cell number quantified (F). Graph shows mean and SEM of the ratios of the cell numbers incubated with and without arginine from 3 independent experiments (*P < .05 by Student’s t-test). (†Separate evaluation of PD-1 expression in Supplementary Figure S7).
Compared to resistant QPP8, sensitive QPP7 showed less PD-L1 induction in the myeloid stroma, particularly following CTLA-4 blockade, but also more consistent upregulation of arginase and TGF-β (LAP) (Figure 5D). CD8 T cells elevated Granzyme B expression in response to both checkpoint antibodies in both tumors, with the only immunologic advantage for QPP7-infiltrating T cells being elevated proliferation in the CD4 effector compartment in response to CTLA-4 blockade. This finding is counterbalanced, however, by the higher expression of both Ki67 (proliferation) and TGF-β (LAP) in Treg from QPP7 versus QPP8. Among these lines, only QPP7 expresses GFP that interfered with PD-1 measurement using our standard flow cytometry panel. In a separate study, we found that PD-1 is downregulated with treatment as in QPP8 (Supplementary Figure S9). To exclude the possibility that differential infiltration of therapeutic antibodies contributes to the immune sensitivity observed in brain QPP7, we determined the in vivo kinetics of the ICB antibodies used in this study. Although there was a higher fluorescent signal indicating more antibody infiltration in brain QPP8 24 h post antibody injection, the level of the antibodies in brain QPP8 became comparable to the levels in brain QPP7 48 h later (Supplementary Figure S10A, B). Ex vivo imaging of the brains harvested 72 h post in vivo injection of antibodies also showed similar antibody levels between QPP7 and QPP8 brains (Supplementary Figure S10C, D). These data suggest that the differential immune sensitivity and immune landscapes in brain QPP7 and QPP8 were not due to the differential penetration of antibodies in different tumors.
To further determine T cell activity, we isolated the intratumoral T cells from the tumors treated with ICB or PBS. We stimulated the intratumoral T cells ex vivo and assessed their IFN-γ, TNF-α, and PD-1 expressions. Consistent with our and others’ previous findings that T cells in human and murine glioblastoma are often exhausted with higher PD-1 expression and limited capacity of inflammatory cytokine production,24,25 we also observed that CD8 T cells and CD4 effector T cells from the ICB-treated QPP7 or QPP8 did not show an elevated IFN-γ or TNF-α expression compared to their PBS treated counterparts (Supplementary Figure S11A, B). Despite the inability of the ICB to enhance effector functions on the intratumoral T cells, the CD8 T cells in the ICB-treated QPP7 but not QPP8 showed a significantly lower PD-1 expression compared to the PBS control, while CD4 T cells also showed a trend of lower expression (Supplementary Figure S11C). The non-reduced PD-1 expression on T cells in QPP8 together with the significant PD-L1 upregulation in the myeloid compartments (Figure 5D) reflect the immune resistance of QPP8. This comparative analysis reinforces the importance of the broad-based upregulation of PD-L1 as an adaptive resistance mechanism for QPP8 but raises questions as to why the arginase upregulation by QPP7 in the brain fails to shield these tumors from checkpoint blockade mobilized T cell responses.The role of arginase in suppressing T cell antitumor immunity has been well established26; however, there are also reports that glioblastoma cells are sensitive to arginine depletion, especially in the context of p53 loss, a shared feature of QPP tumors and most patients.27,28 Administration of arginase as a therapeutic intervention to deplete arginine can limit glioblastoma growth and enhance the efficacy of temozolomide chemotherapy and radiotherapy.27,29 Consistent with prior findings,27 we found that QPP7 and QPP8 undergo proliferative arrest in the absence of arginine in vitro (Figure 5E, F). These data indicate that while upregulation of arginase by QPP7 in response to ICB may cause some inhibition of T cell responses, that deleterious impact on antitumor immunity may be outweighed by the more substantial negative impact on the viability of the QPP tumor cells themselves.
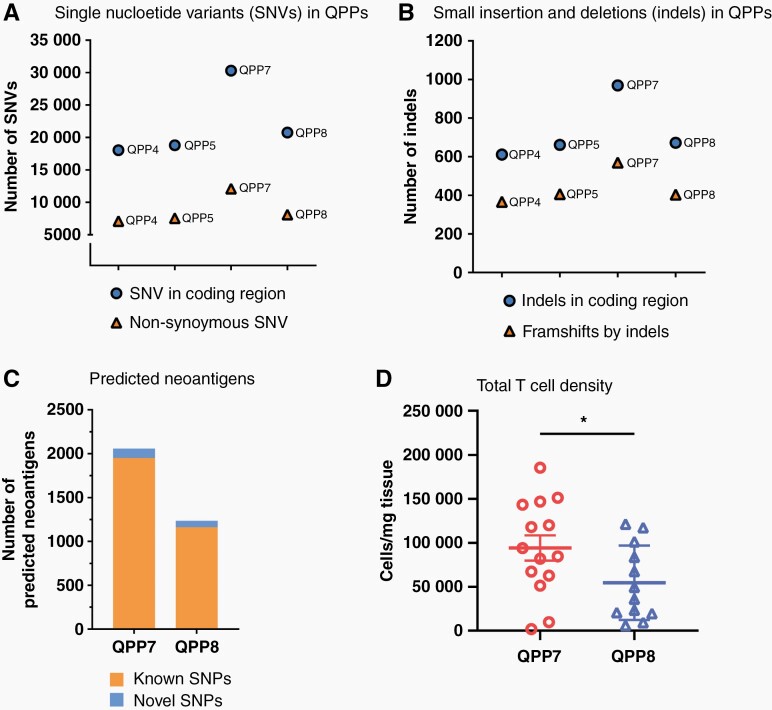
To further clarify the potential mechanisms contributing to the immune sensitivity of QPP7, we evaluated the mutational landscape of these QPP lines. Across immunotherapy, tumors with higher mutational burden generally respond better to checkpoint blockade,30 and pediatric glioma patients with biallelic mismatch repair deficiency are highly responsive to PD-1 blockade.31 As QPP7 was sensitive to any ICB at either site and showed the most evidence for immune editing moving from immune deficient to competent mice, we hypothesized that QPP7 might have a higher antigenicity profile compared to the other lines. Whole exome sequencing and comparative analysis relative to the C57BL/6 reference genome revealed that QPP7 has the highest density of mutations including non-synonymous single-nucleotide variants (Figure 6A), as well as insertions and deletions in coding regions (Figure 6B). The other QPP lines show lesser and relatively homogenous mutational frequencies of ~7.5 mutations/MB (calculated based on exome mutations/ whole genome size) that fall within the normal range for human glioblastoma, but below QPP7 (~ 12 mutations/MB) and well below the frequency for nearly all transplantable mouse models that average 37 mutations/MB.32 QPP7 and GL261 have a similar frequency of single-nucleotide variants in coding regions (30 305 and 26 531, respectively) and a similar number of predicted neoantigens (2058 and 1784, respectively), perhaps explaining their shared sensitivity to anti-PD-1.33 An admitted limitation we faced was lack of available normal tissue from the exact mice from which QPP tumor lines were derived; however, we feel that comparisons to the C57BL/6 reference genome are apt as that is the background in which these lines are implanted. In addition to the quantity of the mutation, increasing literature suggests that the “quality” such as neoantigen load should be considered in predicting the treatment response to immunotherapies.34 Recent studies showed a non-superior response to PD-1 blockade in recurrent glioblastoma patients with a higher mutation burden. Although the higher mutational burden was likely induced by chemotherapy treatment, this finding also suggests the lesser role of using the number of mutational burdens alone in predicting the responses to immunotherapies. Rather, the immune landscape in the TME and the immunogenicity of neoantigen load may play a more important role in the clinical outcome.35,36
Figure 6.
Genomic profile of QPP cell lines. DNA extracted from the cultured QPP lines was sent for whole exome sequencing (Novogene). Tumor mutational burden in each QPP line is presented as the number of single-nucleotide variants (SNV) in coding regions and non-synonymous SNV (A); as well as the number of small insertion and deletion (Indels) and frameshifts by Indels (B). Neoantigens were predicted MuTect and NetMHC program and the numbers of the predicted neoantigens are presented (C). DNA extracted from the CD4 T cells and CD8 T cells isolated from QPP7 and 8 brain tumors were sent for TCR sequencing (ImmunoSeq). TCR richness (D) and Simpson clonality (E) of the intratumoral CD4 and CD8 T cells were calculated and are presented (*P < .05 by Student’s t-test).
In line with the clinical finding, we observed a higher predicted neoantigen load in immune-sensitive QPP7 compared to the immune-resistant QPP8 (Figure 6C). To examine if high neoantigen burdens from the tumor are also associated with higher immunogenicity, we further analyzed the TCR repertoire of the intratumoral CD4 and CD8 T cells isolated from the untreated mice implanted with QPP7 or QPP8. Via TCR sequencing, we observed significantly higher TCR richness of CD4 T cells in the immune-sensitive QPP7 compared to the immune-resistant QPP8 (Figure 6D). Interestingly, TCR clonality of CD4 T cells is not significantly different between QPP7 and 8. TCR clonality of CD8 T cells is significantly higher in QPP8, which suggests a stronger clonal expansion of this subset (Figure 6E). This finding supports the clinical study in which pre-immunotherapy CD4 but not CD8 TCR richness is associated with better outcomes after either anti-CTLA-4 or anti-PD-1 blockades.37 Supported by another pre-clinical study showing that CD4 T cells are necessary for the activation phase of adaptive immune response against cancer,38 these findings highlight the importance of neoantigen load in the tumor and the richness of TCR repertoire of CD4 T cells from the host in predicting checkpoint sensitivity in glioblastoma, even in the context of a suppressive microenvironment.
Discussion
More than 8 years after FDA approval of the first T cell checkpoint antibody, ipilimumab, development of immunotherapy for glioblastoma remains hampered by the lack of pre-clinical tumor models that reflect the immunogenicity of clinical disease. The most commonly used model, GL261, predicts exceptional sensitivity to PD-1 blockade, which generally does not benefit glioblastoma patients.1,6 Genetically engineered mouse models capture key tumor driver genes and can be useful for targeted or chemotherapy evaluations; however, the strength of these mutations often obviates the need for acquisition of secondary mutations leaving an absence of immune-target neoantigens. Xenograft tumor models allow the study of human cancer, but the deficient immune system in the host mice renders them mostly useless for evaluating immunotherapies. Here, we describe QPP cell lines driven by mutations common in human glioblastoma that can be implanted either subcutaneously or intracranially in C57BL/6J mice. The presence of the Qk mutation confers histopathological heterogeneity,10 and differential ICB sensitivity to these tumor lines. Like human glioblastoma, 2 of these lines (QPP4, QPP8) are completely resistant to ICB therapy when growing in the brain. Not only will these new lines be valuable tools to study the origins of immune resistance in glioblastoma as we have done here, but they will also provide critical tools for pre-clinical evaluation of prospective immunotherapies so that only the most promising approaches advance to patients.
Previously, it was shown that high PD-L1 expression predicts poor outcomes in glioblastoma patients.5 PD-L1 is often implicated in “acquired resistance” to immunotherapy in which the IFN-γ produced by T cells elicits upregulation of PD-ligands that dampen the response.39 We also established the capacity of IFN-γ to potently induce PD-L1 on the QPP tumor lines and revealed significantly enhanced PD-L1 expression on the QPP myeloid stroma after ICB, particularly in resistant brain QPP8. Upregulation of PD-L1 in brain QPP8 seemed more pronounced on the myeloid cells than was observed in the more therapeutically sensitive flank site. This may result from differential cytokine and growth factor responses of stroma in each site to checkpoint blockade and could indicate a role for microglia in promoting enhanced PD-L1 induction across the myeloid stroma. These findings illustrate the potential prognostic value of PD-L1 expression by both the tumor cells and the surrounding myeloid stroma. Analysis of the TMEs of QPP8 in the subcutaneous versus orthotopic niches reveals multiple factors associated with treatment resistance. Even in resistant brain QPP8, we observe an increased CD8 T cell frequency in the tumor following ICB; however, we also find signs of acquired resistance. In response to the inflammatory cytokines produced by these T cells, PD-L1 is upregulated across the QPP8 myeloid stroma including on the microglia. In addition to TGF-β and arginase, these cells are known to use PD-L1 to suppress anti-glioblastoma T cells.40 Whereas PD-1 blockade likely prevents induced myeloid PD-L1 adverse impacts in flank QPP8, the PD-1 antibodies fail to rescue T cell function in brain QPP8. This failure could result from inadequate bioavailability in the brain TME, or from other suppressive mechanisms exerting dominant suppression over the release of T cells from PD-1 inhibition.23 These findings are consistent with the key role recently attributed to myeloid cells in human glioblastoma in mediating resistance to ICB.41
QPP7 is the most aggressive tumor line we generated; however, it is also the most immunotherapy sensitive. QPP7 is the only line to respond therapeutically to both CTLA-4 and PD-1 blockade in both the subcutaneous and orthotopic sites. Brain QPP7 showed signatures of a suppressive TME with suboptimal ratios of CD8 T cells versus suppressive stroma, high expression of arginase in the myeloid stroma, and modest induction of PD-L1 in response to ICB. While these adaptations would often be sufficient to grant tumor immune privilege, in this case, they may fail due to a combination of both the adverse impact of arginase on the tumor stem cells and the more diverse antitumor T cell repertoire likely resulting from the higher mutation frequency of QPP7, in addition to its expression of GFP. Although potentially a driver of accelerated tumorigenesis and, the aggressive phenotype of this tumor, this elevated mutation rate and neoantigen load also elicit more diverse and higher affinity antitumor T cell responses such as CD4 TCR richness that may underlie the checkpoint sensitivity we observe.42 Although mutation frequencies are generally low in spontaneous glioblastoma and mismatch repair defects are uncommon,43 studies across many cancers including pediatric gliomas have validated that higher sensitivity to ICB therapy is linked with a higher mutational burden.30,44–46 Despite its relatively low antigen density, systemically administered tumor neoantigen vaccines have proven capable of mobilizing T cells to glioblastoma in the brain.47
In conclusion, we believe that these QPP models provide the most accurate reflection of the immunobiology of human glioblastoma and will contribute to advancing effective immunotherapeutic interventions into clinical practice.
Supplementary Material
Contributor Information
Chao-Hsien Chen, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Neurology, Houston Methodist Neurological Institute, Houston, Texas 77030, USA.
Renee L Chin, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Genevieve P Hartley, Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Spencer T Lea, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Brian J Engel, Departement of Cancer Systems Imaging, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Cheng-En Hsieh, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Rishika Prasad, Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Jason Roszik, Departement of Melanoma Medical Oncology, The University of Texas MD Anderson Cancer Center Houston, Texas 77030, USA.
Takashi Shingu, Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Gregory A Lizee, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Departement of Melanoma Medical Oncology, The University of Texas MD Anderson Cancer Center Houston, Texas 77030, USA.
Amy B Heimberger, Department of Neurosurgery, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Steven W Millward, Departement of Cancer Systems Imaging, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Jian Hu, Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
David S Hong, Department of Investigational Cancer Therapeutics, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Michael A Curran, UTHealth Graduate School of Biomedical Sciences, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA; Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Funding
Dr. Marnie Rose Foundation, The Brockman Foundation, and NCI Cancer Center support grant P30CA16672.
Conflict of interest statement
None related to the content of this manuscript.
Authorship
Experiment design, data analysis, interpretation, and writing manuscript: C.-H.C. and M.A.C.. Tumor growth and immune sensitivity studies: C.-H.C., R.L.C., and S.T.L.. Phenotype characterization and arginine auxotrophy experiments: C.-H.C., R.L.C., S.T.L., and C.-E.H.. Tumor immune infiltrate experiments: C.-H.C., G.P.H., C.-E.H., and R.P.. In vivo imaging of checkpoint blockade antibodies: B.J.E. and S.W.M
Neoantigen prediction: J.R. and G.A.L.. Expert feedback: T.S., J.H., and D.S.H.. Manuscript editing: A.B.H., J.H. and M.A.C.. Study supervision: M.A.C.
References
- 1. Reardon DA, Omuro A, Brandes AA, et al. OS10.3 randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: checkmate 143. Neuro Oncol. 2017;19(Suppl_3):iii21–iii21. [Google Scholar]
- 2. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qazi MA, Vora P, Venugopal C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28(7):1448–1456. [DOI] [PubMed] [Google Scholar]
- 4. Quail DF, Joyce JA.. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nduom EK, Wei J, Yaghi NK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18(2):195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reardon DA, Gokhale PC, Klein SR, et al. Glioblastoma eradication following immune checkpoint blockade in an orthotopic, immunocompetent model. Cancer Immunol Res. 2016;4(2):124–135. [DOI] [PubMed] [Google Scholar]
- 7. Doucette TA, Kong LY, Yang Y, et al. Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro Oncol. 2012;14(9):1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kosaka A, Ohkuri T, Okada H.. Combination of an agonistic anti-CD40 monoclonal antibody and the COX-2 inhibitor celecoxib induces anti-glioma effects by promotion of type-1 immunity in myeloid cells and T-cells. Cancer Immunol Immunother. 2014;63(8):847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Genoud V, Marinari E, Nikolaev SI, et al. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology. 2018;7(12):e1501137e1501137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shingu T, Ho AL, Yuan L, et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat Genet. 2017;49(1):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen AJ, Paik JH, Zhang H, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26(13):1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin D, Ogawa S, Kawamata N, et al. High-resolution genomic copy number profiling of glioblastoma multiforme by single nucleotide polymorphism DNA microarray. Mol Cancer Res. 2009;7(5):665–677. [DOI] [PubMed] [Google Scholar]
- 13. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skelton D, Satake N, Kohn DB.. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther. 2001;8(23):1813–1814. [DOI] [PubMed] [Google Scholar]
- 15. Ager CR, Reilley MJ, Nicholas C, et al. Intratumoral STING activation with t-cell checkpoint modulation generates systemic antitumor immunity. Cancer Immunol Res. 2017;5(8):676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Izumoto S, Ohnishi T, Arita N, et al. Gene expression of neural cell adhesion molecule L1 in malignant gliomas and biological significance of L1 in glioma invasion. Cancer Res. 1996;56(6):1440–1444. [PubMed] [Google Scholar]
- 19. Cheng L, Wu Q, Huang Z, et al. L1CAM regulates DNA damage checkpoint response of glioblastoma stem cells through NBS1. EMBO J. 2011;30(5):800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tchoghandjian A, Baeza N, Colin C, et al. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol. 2010;20(1):211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heiland DH, Haaker G, Delev D, et al. Comprehensive analysis of PD-L1 expression in glioblastoma multiforme. Oncotarget. 2017;8(26):42214–42225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Chaichana KL, Kleinberg L, et al. Glioblastoma recurrence patterns near neural stem cell regions. Radiother Oncol. 2015;116(2):294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. St-Amour I, Pare I, Alata W, et al. Brain bioavailability of human intravenous immunoglobulin and its transport through the murine blood-brain barrier. J Cereb Blood Flow Metab. 2013;33(12):1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ott M, Tomaszowski KH, Marisetty A, et al. Profiling of patients with glioma reveals the dominant immunosuppressive axis is refractory to immune function restoration. JCI Insight. 2020;5(17):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Woroniecka K, Chongsathidkiet P, Rhodin K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mills CD, Shearer J, Evans R, Caldwell MD.. Macrophage arginine metabolism and the inhibition or stimulation of cancer. J Immunol. 1992;149(8):2709–2714. [PubMed] [Google Scholar]
- 27. Hinrichs CN, Ingargiola M, Kaubler T, et al. Arginine deprivation therapy: putative strategy to eradicate glioblastoma cells by radiosensitization. Mol Cancer Ther. 2018;17(2):393–406. [DOI] [PubMed] [Google Scholar]
- 28. Pavlyk I, Rzhepetskyy Y, Jagielski AK, et al. Arginine deprivation affects glioblastoma cell adhesion, invasiveness and actin cytoskeleton organization by impairment of beta-actin arginylation. Amino Acids. 2015;47(1):199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Przystal JM, Hajji N, Khozoie C, et al. Efficacy of arginine depletion by ADI-PEG20 in an intracranial model of GBM. Cell Death Dis. 2018;9(12):1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yarchoan M, Hopkins A, Jaffee EM.. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 32. Zeitouni B, Tschuch C, Davis JM, et al. Abstract 1840: whole-exome somatic mutation analysis of mouse cancer models and implications for preclinical immunomodulatory drug development. Cancer Res. 2017;77(13_Suppl):1840–1840. [Google Scholar]
- 33. Johanns TM, Ward JP, Miller CA, et al. Endogenous neoantigen-specific cd8 t cells identified in two glioblastoma models using a cancer immunogenomics approach. Cancer Immunol Res. 2016;4(12):1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strickler JH, Hanks BA, Khasraw M.. Tumor mutational burden as a predictor of immunotherapy response: is more always better? Clin Cancer Res. 2021;27(5):1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gromeier M, Brown MC, Zhang G, et al. Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat Commun. 2021;12(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arakawa A, Vollmer S, Tietze J, et al. Clonality of CD4(+) blood T cells predicts longer survival with CTLA4 or PD-1 checkpoint inhibition in advanced melanoma. Front Immunol. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zander R, Schauder D, Xin G, et al. CD4(+) T cell help is required for the formation of a cytolytic CD8(+) T cell subset that protects against chronic infection and cancer. Immunity. 2019;51(6):1028–1042.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A.. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hambardzumyan D, Gutmann DH, Kettenmann H.. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Groot J, Penas-Prado M, Alfaro-Munoz KD, et al. Window-of-opportunity clinical trial of pembrolizumab in patients with recurrent glioblastoma reveals predominance of immune-suppressive macrophages. Neuro Oncol. 2019;22(4):539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee CH, Yelensky R, Jooss K, Chan TA.. Update on tumor neoantigens and their utility: why it is good to be different. Trends Immunol. 2018;39(7):536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hodges TR, Ott M, Xiu J, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19(8):1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.