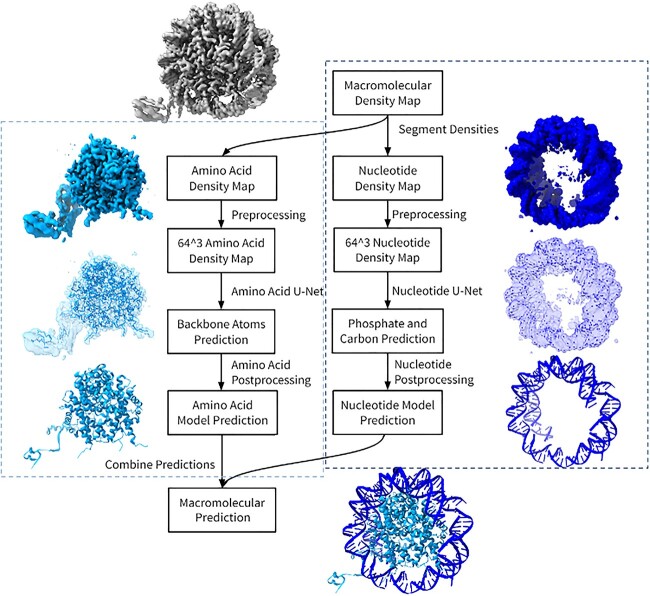
Figure 3.
The system design of the macromolecular pipeline. The pipeline is organized into four major steps: segmentation, preprocessing, their respective machine learning and postprocessing steps. Both amino acids and nucleotides share the same segmentation and preprocessing of their density maps. Once the cryo-EM maps values are normalized and resized to fit a 64 [3] shape, the amino acid U-Net determines the positions of the Cα and other backbone atoms, while the nucleotide U-Net determines the position of phosphate atoms and sugar carbon atoms. Both outputs will be processed through separate postprocessing steps to add on atoms and complete a structure. The models are then combined to generate a complete macromolecular structure. The cyan indicates DeepTracer-1.0’s amino acid pipeline and the dark blue are DeepTracer-2.0’s nucleotide pipeline.