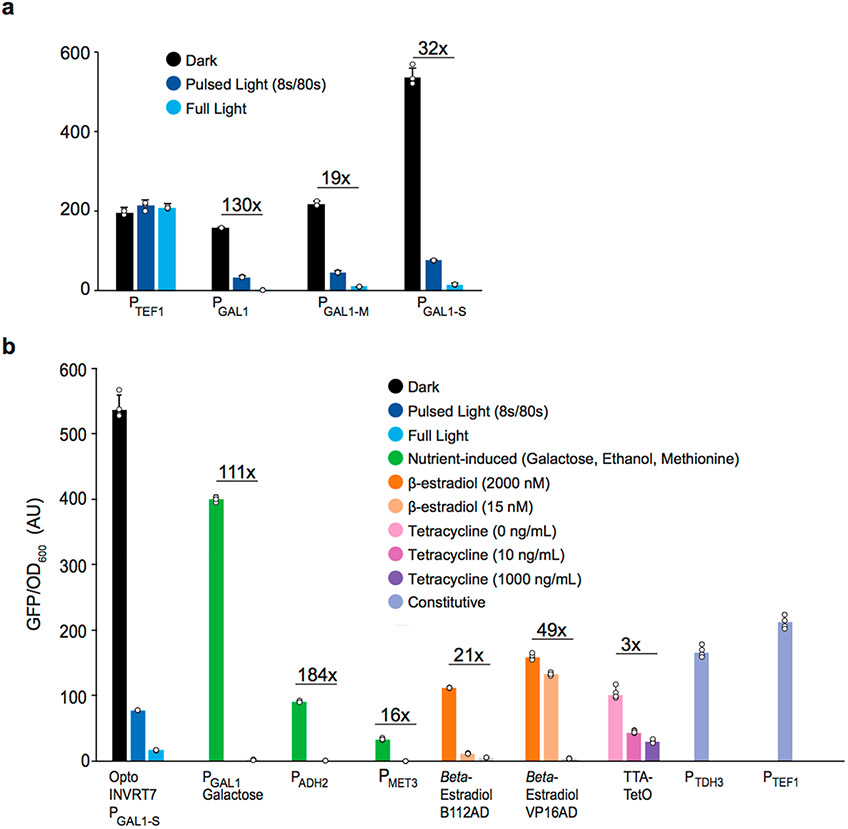
Figure 2.
PGAL1 promoter engineering. (a) Specific GFP expression by engineered GAL1 promoters controlled by OptoINVRT7: PGAL1 (YEZ230C), PGAL1-M (YEZ230) with deleted Mig1p binding sites; PGAL1-S (YEZ229) with four extra Gal4p-binding added to the PGAL1-M sequence. (b) Specific GFP expression controlled: optogenetically (blue and black) by darkness-induced OptoINVRT7 (YEZ229); with nutrients (green): galactose-induced PGAL1 (YEZ48), ethanol-induced PADH2 (YEZ294) and methionine-repressed PMET3 (YEZ295); with synthetic promoters: β-estradiol-induced (orange) LexABD-ER-B112AD31 (yMAL34), or LexABD-ER-VP16AD31 (yMAL35), and tetracycline-repressed (pink and purple) tTA32,33 (yMAL36); or constitutive promoters (periwinkle): PTDH3 (YEZ228) and PTEF1 (YEZ186). Light-sensitive strains in (a) and (b) are shown in full light (light blue), 8 s ON/72 s OFF (dark blue), and full darkness (black); yMAL34 and yMAL35 in 2000 nM, 15 nM, and 0 nM β-estradiol31 and yMAL36 in 0, 10, and 1000 ng/mL tetracycline.33 YEZ140 (no GFP control) was used to subtract background and autofluorescence. All data are shown as mean values; dots represent individual data points; error bars represent the s.d. of four biologically independent 1-mL sample replicates exposed to the same conditions. All experiments were repeated at least three times.