Fig. 1.
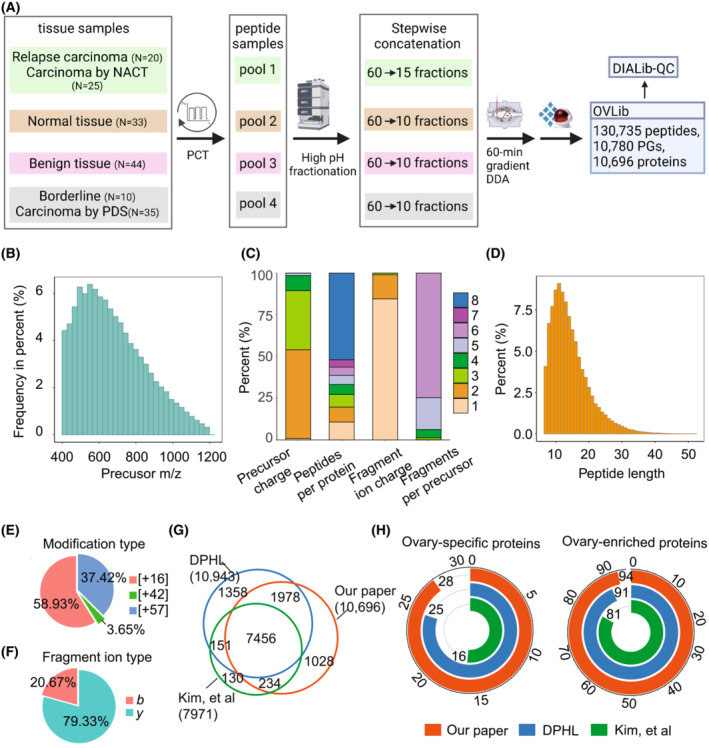
The ovary spectral library. (A) Workflow for the generation of an OVLib. PGs represent protein groups. (B) The distribution of precursor m/z. NACT, Neoadjuvant chemotherapy; PDS, Primary debulking surgery; PCT, Pressure cycling technique; DDA, data‐dependent acquisition; OVLib, Ovary spectral library; DIALib‐QC, DIA library quality control; PGs, protein groups. (C) The distribution of precursor charge states, proteotypic peptides for each protein, fragment ions per precursor ion, and fragment ion charge states. m/z, Mass‐to‐charge ratio. (D) The distribution of peptide lengths. (E) The distribution of three modifications (16: oxidation in methionine, 42: acetylation in N terminal, 47: carbamidomethylation in cysteine). (F) The proportion of b, y ions. (G) The Venn plot of unique proteins among OVLib and two prepublished libraries. DPHL, DIA pan‐human spectral library. The counts of ovary‐specific proteins and ovary‐enriched proteins (H) among OVLib and two prepublished libraries.
