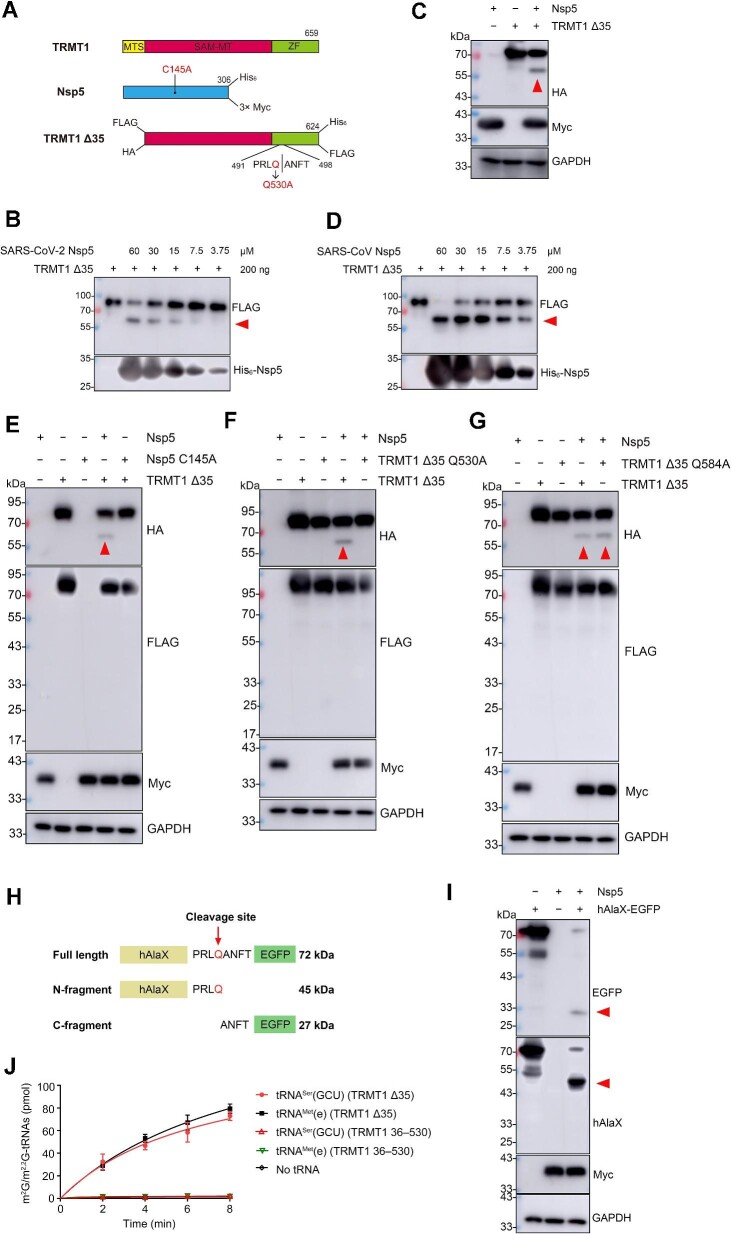
Figure 1.
Nsp5 cleaves and inactivates TMRT1 in vitro and in vivo. (A) Diagram of the expression constructs of TRMT1, TRMT1 Δ35, Nsp5, and mutated forms of TRMT1 Δ35 and Nsp5. The SAM methyltransferase (SAM-MT) domain and the zinc finger (ZF) domain were shown in red and green, respectively. For each recombinant protein, tags labelled in the prokaryotic expression system are shown on the top, while tags labelled in the eukaryotic expression system are shown on the bottom. (B) Immunoblotting analysis of the cleavage product of TRMT1 Δ35 by SARS-CoV-2 Nsp5 in vitro. The amount of TRMT1 Δ35 and different concentrations of Nsp5 are indicated. (C) Cleavage of TRMT1 Δ35 by Nsp5 in HEK293T was detected by western blot analysis. (D) Cleavage of TRMT1 Δ35 by SARS-CoV Nsp5 in vitro was detected by western blot analysis. (E) Cleavage of TRMT1 Δ35 by Nsp5 or its inactive mutant (C145A) in HEK293T was detected by western blot analysis. (F and G) Cleavage of TRMT1 Δ35 and its mutants (Q530A and Q584A) by Nsp5 in HEK293T was detected by western blot analysis. (H) Schematic representation of the hAlaX-EGFP reporter with the predicated cleavage site from TRMT1. (I) Cleavage of hAlaX-EGFP by Nsp5 in HEK293T was detected by western blot analysis using anti-hAlaX or anti-EGFP antibodies. (J) Time course curves of m2G/m2,2G modification levels of tRNAMet(e) and tRNASer(GCU) catalyzed by TRMT1 Δ35 and TRMT1 36–530. The data represented mean ± standard deviation (n = 3). In all relevant images, the red arrow indicates the digested product of TRMT1 Δ35 or hAlaX-EGFP.