Figure 2: Generated artificial antibacterial proteins are diverse and express well in our experimental system.
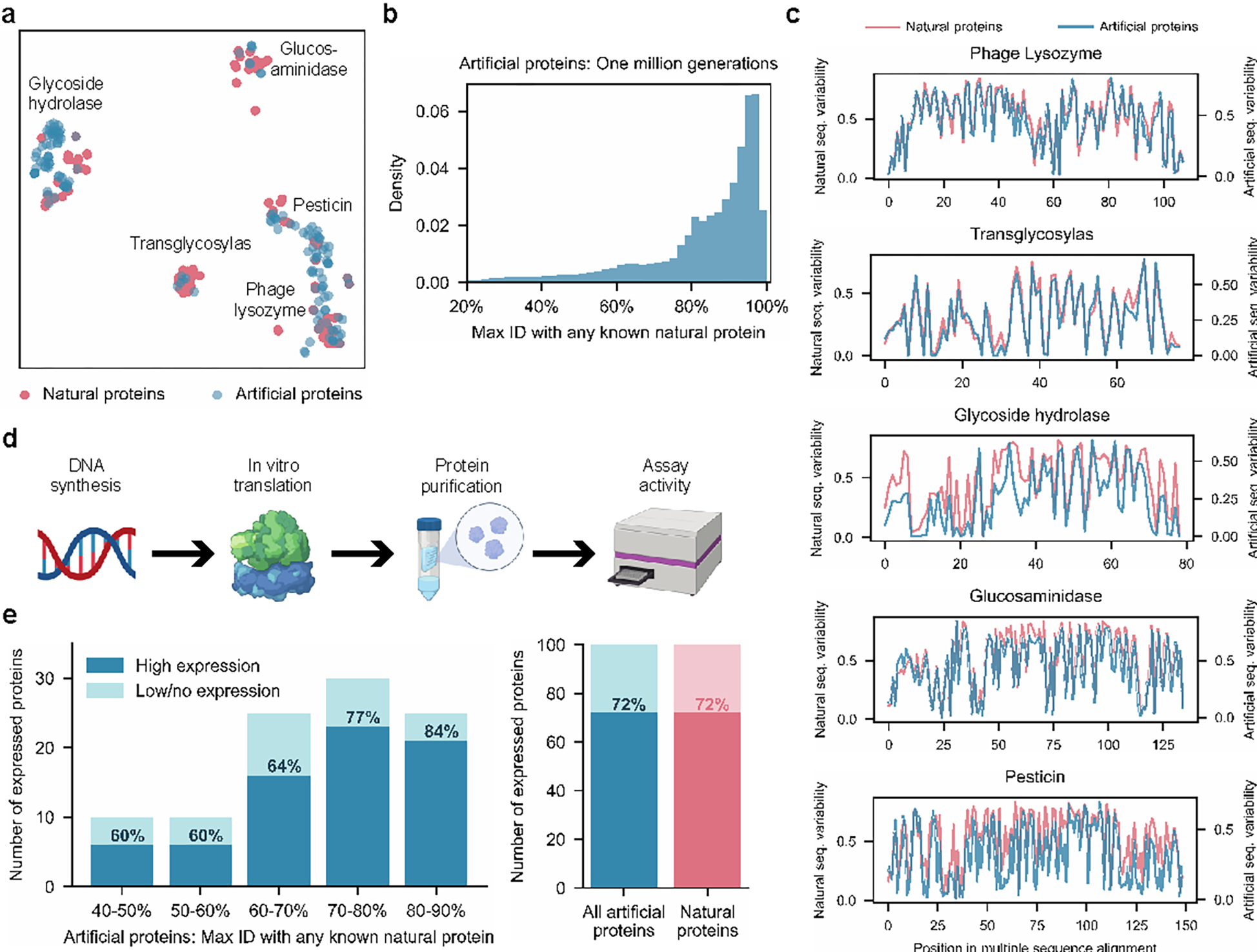
When analyzed using t-SNE as a dimensionality reduction technique for visualization purposes, artificial sequences from our model are shown to span the landscape of natural proteins from five lysozyme families (a). Each point represents a natural or generated sequence embedded in a two-dimensional t-SNE space. With sufficient sampling, ProGen can generate sequences that are highly dissimilar from natural proteins (b). Max ID measures the maximum identity of an artificial protein with any publicly available natural protein. (c) Artificial proteins maintain similar evolutionary conservation patterns as natural proteins across families. Plots demonstrate the variability at each aligned position for a library of proteins. Conserved positions are represented as curve dips. From our generated proteins, we select one hundred proteins for synthesis and characterization in our experimental setup (d). Artificial proteins express well even with increasing dissimilarity from nature (40–50% max ID) and yield comparable expression quality to one hundred representative natural proteins (e).
