Figure 3: Artificial protein sequences are functional while reaching as low as 44% identity to any known protein, exhibit comparable catalytic efficiencies to a highly-evolved natural protein, and demonstrate similar structures to known natural folds.
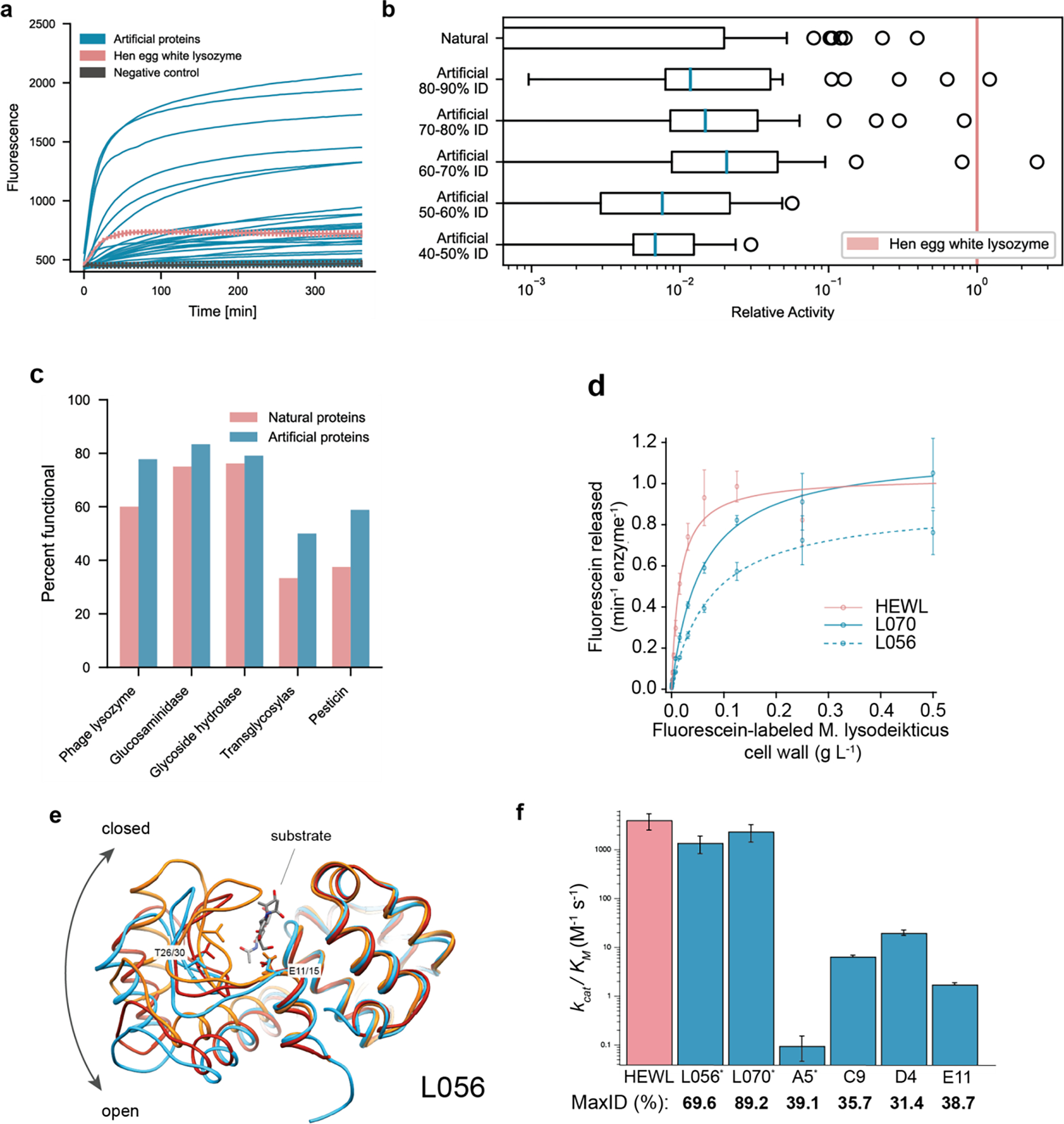
(a) Artificial proteins bind well to substrates and exhibit high fluorescence responses over time. Error bars (minimum and maximum) are shown for hen egg white lysozyme, HEWL, and negative (ubiquitin) controls. (b) Artificial proteins remain active even while being dissimilar (40–50% max ID i.e., top hit-identity) from known natural proteins. Outliers indicate high activity samples where relative activity is computed with respect to HEWL. (c) Artificial proteins are functional across protein families. Functional is defined as a fluorescence one standard deviation above the maximum value of all negative controls. (d) Michaelis-Menten kinetics of HEWL natural lysozyme (red) and two generated lysozymes (blue; L056 and L070) against cell-wall substrate show comparable performance ( technical replicates). (e) We determined a 2.5 Å resolution crystal of L056 artificial lysozyme. A global overlay of L056 crystal structure with two representative T4 lysozyme conformations is shown with L056 presented in sky blue, ‘open’ conformation of M6I T4 lysozyme (PDB:150L) in dark red, ‘closed’ conformation of wild-type T4 lysozyme (PDB:3FA0) in orange, and substrate (PDB:148L) colored by element. Catalytic threonine (T30 in L056 and T26 in T4 lysozyme) and first catalytic glutamate (E15 in L056 and E11 in T4 lysozyme) are represented as sticks. (f) Michaelis-Menten constants derived for lysozyme variants demonstrating a range of catalytic activities across variants of varied maximal sequence IDs to known natural protein. Error bars represent propagated standard deviations (). * denotes derived from initial rate analysis and unit converted (Table S4).
