Figure 4: Applicability of conditional language modeling to other protein systems.
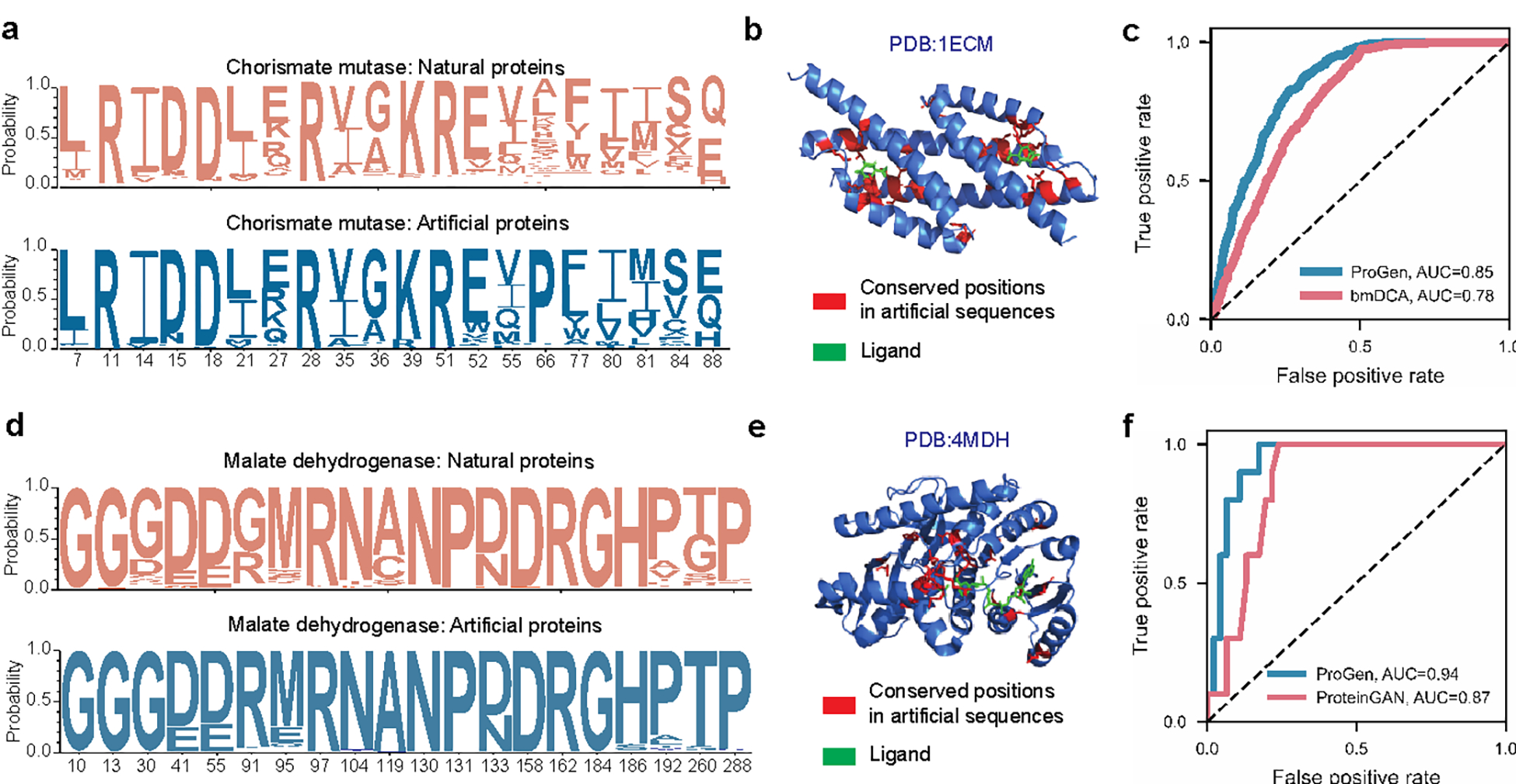
Using the appropriate control tag, our language model, ProGen, can generate sequences for distinct protein families. Here we show that ProGen can generate chorismate mutase (CM) enzymes that exhibit a similar residue distribution to nature (a) and the conserved residues among generated sequences correlate to ligand-binding sites (b). ProGen’s model likelihoods can also accurately predict the functionality of CM variants from published data, slightly better than the coevolutionary bmDCA7 algorithm from the original study (c). ProGen can also generate malate dehydrogenase (MDH) proteins that exhibit a similar residue distribution to nature (d). The conserved residues among generated sequences correlate to buried residues (e). ProGen’s model likelihoods are also accurate in predicting functionality of published variants of MDH, similar to the generative proteinGAN66 model used in the original study (f).
