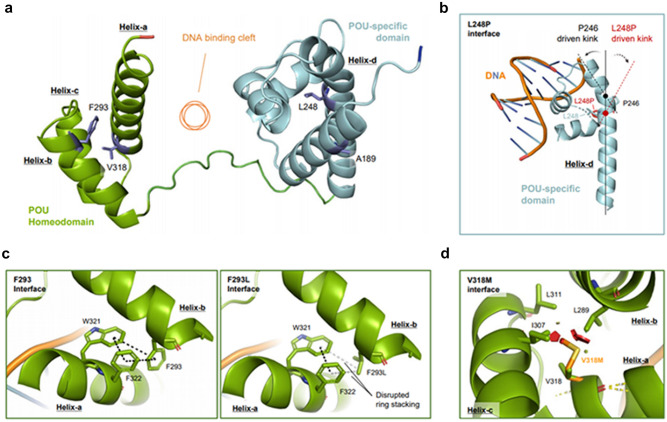
Figure 3.
Novel POU4F3 variants destabilize the inter-helical interactions, impairing the transcriptional activity of POU4F3. (a) Sideview of Alphafold generated model structure of POU4F3. (Jumper et al.37 Highly accurate protein structure prediction with AlphaFold., Nature). POU homeodomain (green) and POU-specific domain (cyan) assembled with DNA binding cleft (orange circle) in between. Val318 and Phe293 are present in the Helix-a and Helix-b of the homeodomain (green), respectively, while Leu248 is in the Helix-d of the POU-specific domain (cyan). All the mutant residues are facing intra-helical spaces, not directly interacting with DNA. (b) Intra-helical proline substitution at Leu248 causes helical kinks. A 27-amino acid long helix-d has a natural kink (black dotted line) driven by Pro246 in the middle. Additional proline substitution induces the formation of an additional kink (red dotted line) starting from p.Leu248Pro (red dot), causing dramatic conformational changes in the POU-specific domain. (c) Phe293 forms aromatic ring stacking (black dotted line) with Trp321 and Phe322 in helix-a (left), stabilizing the interhelical interface. The p.Phe293Leu variant largely disrupts biochemical interactions between helix-a and helix-b, destabilizing helical assembly of POU-homeodomain. (d) Key amino acid residues of intramolecular hydrophobic cavity of POU homeodomain. Val318 forms hydrophobic interactions with Ile307, Leu289, and Leu311. The Val318Met mutant with long side chain clashes (red polygons), with the adjacent Ile307 and Leu289, changing the distance between helices. L Leu, V Val, P Pro, F Phe, W Trp, I Ile, M Met.