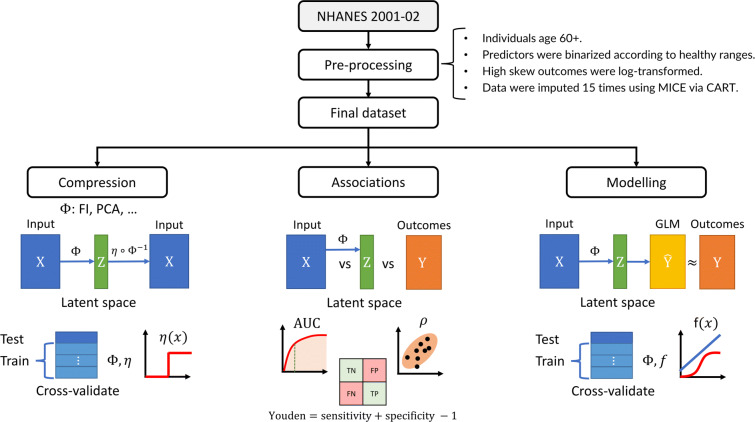
Fig. 1.
Study pipeline. We performed three parallel analyses: compression, feature associations, and outcome modelling. Data were preprocessed, resulting in an input matrix of health deficit data, X, and an outcome matrix of adverse outcomes, Y (rows: individuals, columns: variables). The input was transformed by a dimensionality reduction algorithm, represented by Φ, which was either the FI (frailty index), PCA (principal component analysis), LPCA (logistic PCA), or LSVD (logistic singular value decomposition). Each algorithm, Φ, generated a matrix of latent features with tunable dimension, Z (dimension: number of columns/features; the FI was not tunable). We tuned the size of this latent feature space, Z, to infer compression efficiency and the maximum dimensions of Z before features became redundant (binarizing with optimal threshold, η). The latent features were then associated with input and outcomes to infer their information content and the flow of information from input to output. The dimension of Z was then again tuned to predict the adverse outcomes. represents the outcome estimates by the generalized linear model (GLM), which were compared to ground truth, Y, to determine the minimum dimension of Z needed to achieve optimal prediction performance for each outcome. This procedure allowed us to characterize the flow of information through each dimensionality reduction algorithm