Fig. 1.
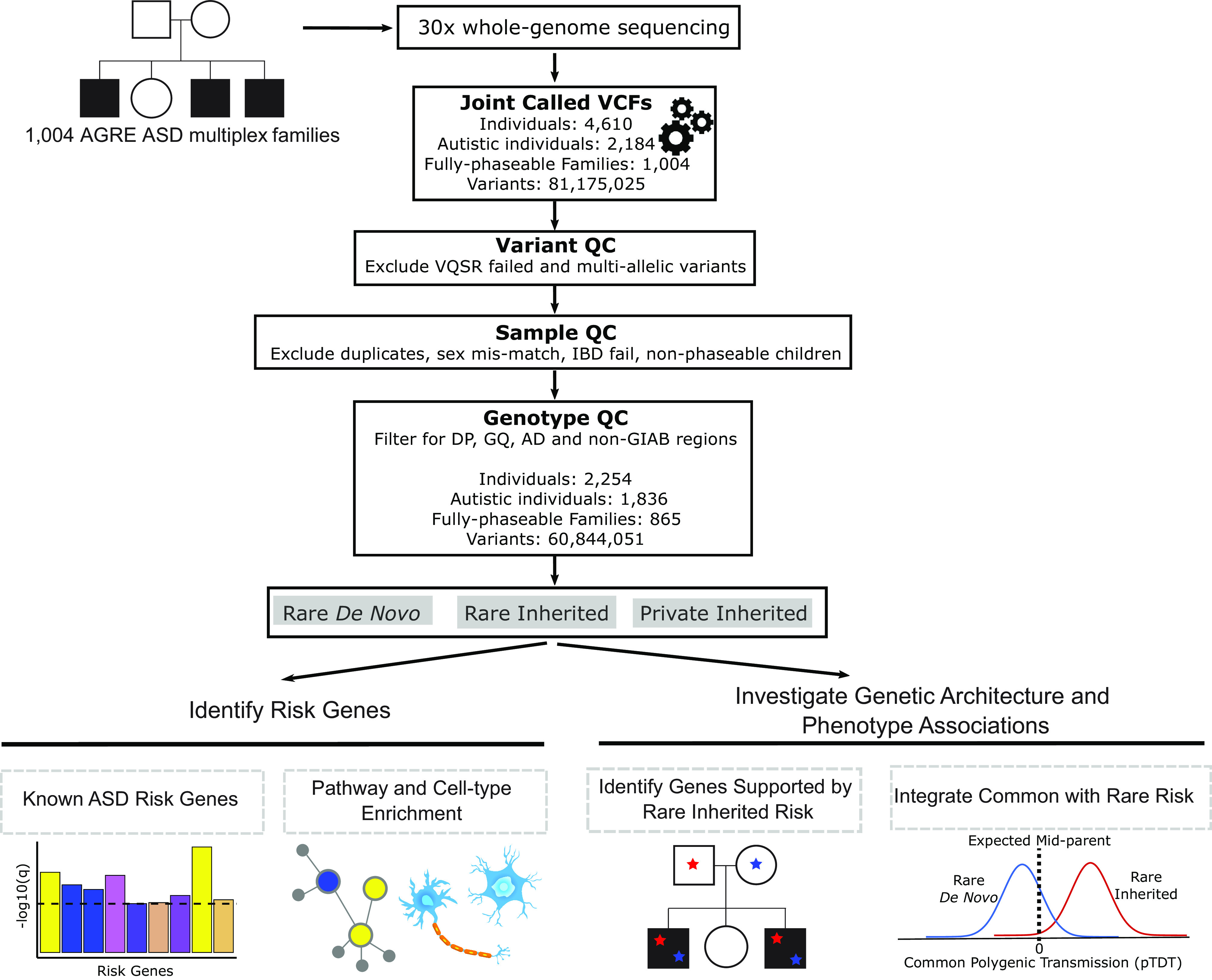
Overview diagram of study analyses. WGS was performed on 1,004 AGRE ASD multiplex families. Single-nucleotide variants and small insertions/deletions were called using the Genome Analysis Toolkit. Variant, sample, and genotype quality control was performed. Rare de novo, rare inherited, and private inherited variants were included for downstream analyses. We identified ASD risk genes at FDR < 0.1 using the Transmission and De Novo Association (TADA) analysis. Risk genes were characterized using integrative genomics. We investigated known genes supported by rare inherited risk and evaluated their variants’ coinheritance and effect on phenotypic measures. We tested if ASD polygenic score was overtransmitted to autistic and nonautistic children stratified by carrier status for rare de novo and inherited variants. VCF = Variant Call Format, QC = Quality Control, VQSR = Variant Quality Score Recalibration, IBD = Identity By Descent, DP = Total Depth, GQ = Genotype Quality, AD = Allele Depth, GIAB = Genome In A Bottle.
