Significance
How complex ecosystems arise and persist has long been a central ecological issue. Attempts to answer this question have typically proceeded by looking at how the percentage of stable assemblages, randomly generated, scales with diversity. Many results generated this way suggest that large stable systems are rarely found in models, in seeming contradiction to the common observation of large complex systems in nature. Here, instead, we use a new inverse approach by assuming that complex systems do exist and characterizing the interactions that would produce this outcome. Finding the kinds of interactions that do produce large complex systems has important implications both for understanding fundamental ecological questions and for understanding the impact of anthropogenic influences on ecological systems.
Keywords: food webs, stability, inverse problems, diversity, network reconstruction
Abstract
Ecologists have long sought to understand how diversity and structure mediate the stability of whole ecosystems. For high-diversity food webs, the interactions between species are typically represented using matrices with randomly chosen interaction strengths. Unfortunately, this procedure tends to produce ecological systems with no underlying equilibrium solution, and so ecological inferences from this approach may be biased by nonbiological outcomes. Using recent computationally efficient methodological advances from metabolic networks, we employ for the first time an inverse approach to diversity–stability research. We compare classical random interaction matrices of realistic food web topology (hereafter the classical model) to feasible, biologically constrained, webs produced using the inverse approach. We show that an energetically constrained feasible model yields a far higher proportion of stable high-diversity webs than the classical random matrix approach. When we examine the energetically constrained interaction strength distributions of these matrix models, we find that although these diverse webs have consistent negative self-regulation, they do not require strong self-regulation to persist. These energetically constrained diverse webs instead show an increasing preponderance of weak interactions that are known to increase local stability. Further examination shows that some of these weak interactions naturally appear to arise in the model food webs from a constraint-generated realistic generalist–specialist trade-off, whereby generalist predators have weaker interactions than more specialized species. Additionally, the inverse technique we present here has enormous promise for understanding the role of the biological structure behind stable high-diversity webs and for linking empirical data to the theory.
Nature is replete with vast, complex networks of interactions that have proven remarkably persistent on ecological timescales (1) but are now threatened by global change (2). The reticulate yet persistent aspect of food webs has led to the search for underlying biological structures that lead to resilience. Arguably, now more than ever, the identification of fundamental biological structures that augment the maintenance of diversity is critical to uncover. Global change is leading to not only the loss of diversity but the wholesale rewiring of complex food webs (e.g., refs. 3–5).
Drawing from a similar methodology applied to complex economic systems (6, 7), Robert May famously introduced the community matrix as a technique to study theoretically the role of diversity and complexity in ecosystem stability (8). The power of these approaches is that they facilitate analysis by employing an enormously simplified mathematical technique that encodes all interactions from nature’s entangled web in a single matrix. This power comes at a cost, though, and the technique has long been criticized for its underlying simplifying assumptions (e.g., May’s statistical universe) that often omit the natural constraints that must exist in empirical webs (9–12). One of the most fundamental of these natural constraints that is omitted is that the randomly generated matrices, in their most general form, do not require feasibility [i.e., that the matrices represent communities of positive biomass; (13, 14)]. Further, frequently, these same matrices make no attempt at constraining interaction strengths to follow known energetic properties of real systems (e.g., where consumers have imperfect conversion of energy from their food sources).
Researchers have historically and recently looked into the role of feasibility and biological structure on the stability of diverse ecosystems (10, 15, 16). One approach has expanded simple dynamical model approaches to more complex bioenergetically realistic dynamical models (e.g., refs. 10 and 17). The dynamical model approach finds that energetic structures that come from the ubiquitous allometric properties of organisms play a key role in stabilization, through the modification of interaction strength often skews them toward weaker interaction strength distributions with fewer strong interactions (e.g., refs. 17 and 18). A second approach has also arisen that implements feasibility into the complex matrix models that Robert May started (16). These approaches often make elegant simplifying assumptions on the matrices to enable analytical solutions (16). Here, researchers have found that feasible webs are related to the number of interactions and the mean strength of interactions (16). Interested in the role of energetically constrained models, recent matrix models without feasibility constraints have also looked at the empirical asymmetry in interaction strengths (i.e., predator growth weaker than prey loss) finding that they tend to produce results entirely consistent with dynamic modular theory (19). Specifically, strong interactions are inherently destabilizing unlike the symmetric predator–prey interaction strength assumption (19).
Nonetheless, the role these biological constraints play in complex communities remain not fully understood. Most work considers a single constraint at a time and the need for approaches that can clearly and easily add multiple biological constraints are needed. Additionally, the elegant simplifications of recent matrix approaches (16, 20) would benefit from less simplified approaches that explore the set of all possible answers. This latter aspect of theory remains computationally intensive as diverse simulated matrices of realistic topology with equilibrium structure are enormously computationally taxing. For example, food web theory currently does not have algorithms to efficiently do highly replicated high-diversity webs of known topologies. While inverse methods in ecology abound and are already found in food web theory to estimate energy flux through webs, they tend to employ many constraints to reduce the complexity of the model and make it computationally more tractable (21, 22). Applied work like EcoPath, for example, often adds empirically motivated constraints to the point that a single solution can be calculated, while more recent methods like the linear inverse method (LIM) have looked at less constrained assumptions to generate distributions of likely flows (22).
Our work here is more consistent with the LIM approach and proceeds by reducing all biological constraints to two core assumptions i) a realistic topology and; ii) feasibility of a known equilibrium solution. Here, we develop a feasibility diversity–stability approach that can serially add in known realistic biological constraints one at a time (e.g., energetic asymmetry between predator and prey) to fully, and clearly, elucidate the role of any given constraint on the local stability of complex diverse food webs. Here, we do this for consumptive predator–prey webs although it could be done more generally for all interaction types. In what follows, we proceed by adding in energetic constraints (asymmetric strengths in predator–prey pairs) to feasible, topologically realistic webs as energetic constraints have played a large role in understanding complex community dynamics (10, 17).
Recently, bioinformaticians interested in the structure and functioning of genomic metabolic networks have come up against a high-dimensional problem with a similar mathematical structure (23, 24). Here, they start off with a given set of steady-state metabolite levels and use these relatively precise empirical estimates to obtain the parameters of the system. Intriguingly, they employ a method that inverts the typical approach used to study community matrices. The theoretical approach starts by specifying the state of the system, the steady-state metabolite levels, and then solving for the system that would produce these levels. This inverse-metabolic network method suggests a similar approach to the community matrix problem. That is, akin to the observations of steady-state metabolite levels, ecologists often have a better idea of the biomass densities (e.g., Eltonian biomass pyramid) than the matrix parameters governing flux rates between nodes. Further, and intriguingly, genomics researchers are also interested in adding biological constraints (e.g., on fluxes) to hone the estimates of the network parameters. This immediately suggests that the same inverse numerical techniques, with the potential for added biological constraints, can be employed for the community matrix. Thus, in the same way, the late Robert May drew from another discipline to attack diversity stability relationships in ecology, below we employ the inverse method with the same computational machinery developed by genomics researchers.
In what follows, we first briefly outline the inverse methodology and its meaning for a community matrix. We then show that this technique in diversity–stability applications allows us to solve the feasibility problem more rapidly while easily adding further biologically plausible constraints (e.g., energetic constraints on interaction; bottom-heavy versus top-heavy food webs [sensu (25)]. We then reexamine longstanding diversity–stability relationships starting with the purely random classical model with realistic topological structure, before adding to this base model a feasibility constraint, and finally including feasibility plus energetic constraints (i.e., consumer gains less energy than it consumes). Importantly, the inverse method uses the biological constraints to filter out interaction strengths, leaving behind interaction strength structures that drive feasibility and stability. Our results show that including feasibility and energetic constraints significantly alter interaction strength structure and increase the likelihood of stability in diverse webs relative to more traditional matrix approaches. This stability can be shown to come from different combinations of interaction strength structures in the diagonal (self-regulation) and off-diagonal terms (species interaction strengths). Specifically, for the classical matrix and the feasibility matrix—consistent with other work (12, 26)—we find that strong self-regulation is a requirement for stable diverse webs. However, with the addition of biological realistic energetic constraints, stable webs arise even with weak self-regulation but appear to require the matrices to be dominated by weak species interactions. We then use the inverse approach to find where these weak interactions manifest. The biologically constrained matrices reveal a naturally arising generalist–specialist trade-off whereby generalist predators have weaker interactions than more specialized species. We end by emphasizing that this inverse technique has enormous promise for further advances in diversity–stability theory and linking this theory to empirical data.
The Inverse Problem: Harnessing the Power of Biological Information
Here, we briefly review the well-known classical diversity–stability matrix approach to highlight an inverse approach—effectively we rephrase the question as an inverse problem in a very straightforward way. We argue that the inverse approach allows us to harness data that are more attainable empirically and the reason related inverse approaches exist in food web research and applications (21). We begin with the typical Lotka–Volterra models used to describe species interactions:
| [1] |
where is the diversity of the system, is the intrinsic growth rate of species , and is the interaction coefficient for species on species .
The classical matrix approach has been to assign the interaction coefficients according to some rule, often randomly, and then to look at the resulting system for equilibria and stability (27–29). Given interaction terms, the system (Eq. 1) can be solved for equilibrium abundances ( ), but unfortunately for random biologically unconstrained parameters, it is unlikely that almost any equilibrium value will be feasible (30).
Following its name, the inverse approach rephrases the problem by instead starting with the assumption that the species abundances ( ) are known. While estimates of density are not trivial, empirical estimates of species density are far better quantified than the enormity of information required for interaction strength data. Further, given that there is additional basic information about intrinsic growth rates of species ( ), this assumption can be biologically constrained even further. These sets of assumptions literally turn upside the classical assumptions and in doing so are more consistent with what we know as biologists.
With these assumptions, we now rephrase the mathematics such that the abundances, , the network structure, and estimates of the growth rates act as constraints on the possible interactions . We then solve for the interaction coefficients instead of the abundances. This rephrasing leads to a severely underdetermined system with many more unknowns than equations (the so-called Inverse Problem). The underdetermined aspect of this leads to a set of possible answers. It is worth pointing out that even in a high-diversity web (i.e., high-dimensional problem), the new rephrasing is a more solvable problem. Indeed, this is a problem that is also shared in bioinformatics where a general numerically intensive algorithm has been developed based on the “Hit and Run” method (31, 32). In a sense, the biological constraints (e.g., equilibrium observed densities) used to construct the inverse problem act as a filter on the interaction strength structure allowing us to clearly see the role interaction strength plays in producing feasible high-diversity webs. For clarity, we first illustrate this approach using the familiar consumer–resource model (schematically represented in Fig. 1, 1–5) before discussing how this is easily extended to high-diversity problems.
Fig. 1.
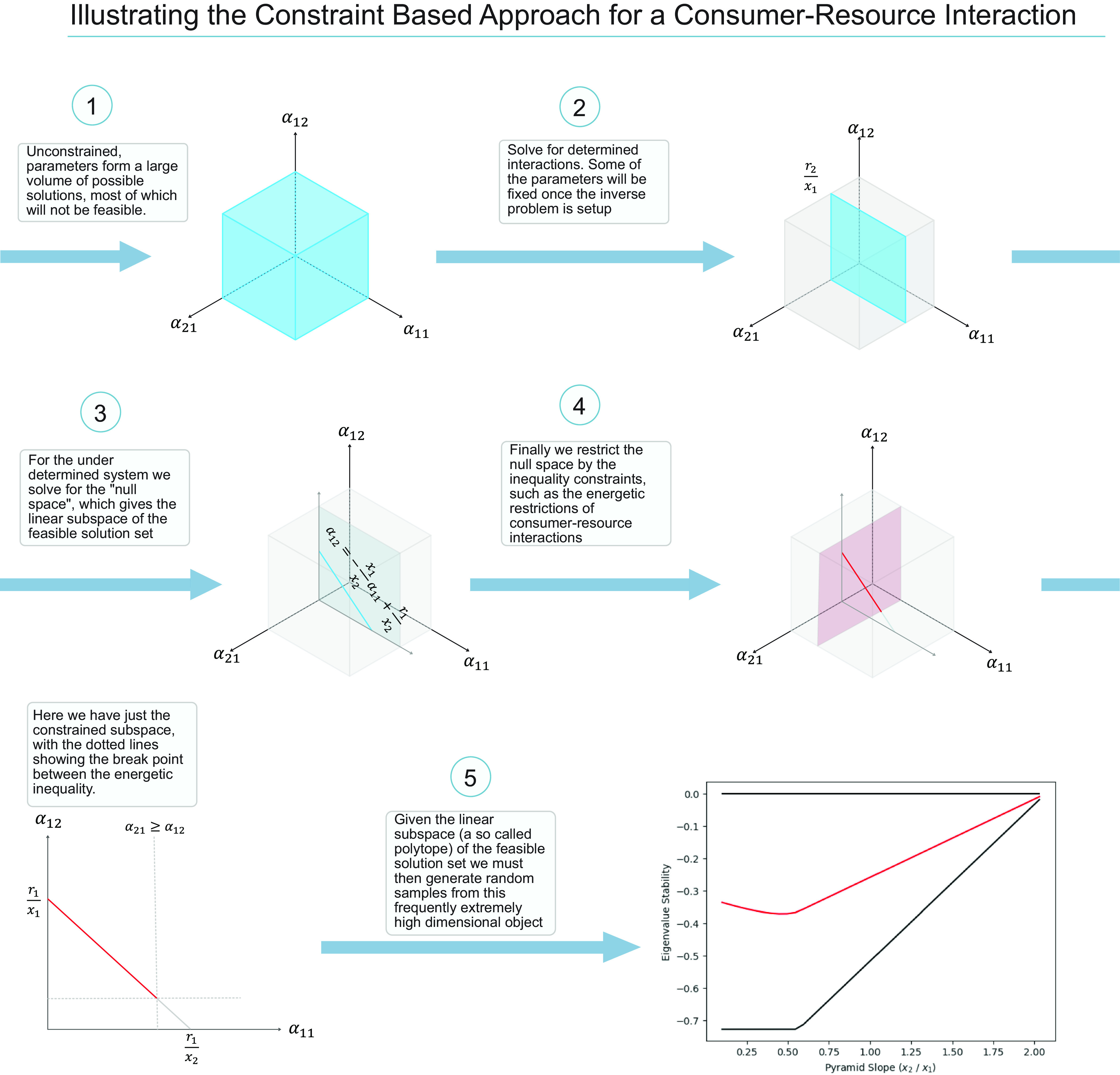
Overview of the mathematical approach used here that bridges the classical approach (1) to the inverse method under different constraints [just feasibility (3) and feasibility with an energetic constraint (5)] for the Lotka–Volterra consumer resource model. Circles 1–5 show the methodology of this paper which moves from the classical random model (1) via the inverse approach to feasible biological constrained models (5). In panel 5, the red line is the mean response of all runs when you are uniformly sampling from the blue line in panel 4, and the dark solid lines are the theoretical minimum and maximum stability responses (i.e., determined from the geometry in panel 4). The numerical values for panel 5 are r = [1.5, −0.5], x1 = 1, x2 in [1/3, 3].
A Concrete Example: The Inverse Method for Consumer–Resource Interactions.
For a classical Lotka–Volterra consumer–resource model, there are two dynamic equations (for x2 and x1; classically C and R for consumer and resource) and three interaction coefficients (the per capital impact of the basal resource on itself, ); the negative per capita impact of the consumer on the resource, ; and the positive benefit the resource gives to the consumer from consumption, ).
From the classical methodological perspective, we seek a solution of the following system:
| [2] |
where biomass equilibrium values (xi) satisfy the Matrix equation with A the matrix of interaction coefficents: . The term indicates that the consumer has a mortality rate, not a growth rate like the resource (14). The classical approach has been well described and sets up conditions for a “statistical universe” of possibilities as exemplified in Fig. 1, 1 (box of solutions constrained only by the parameters assumptions). As discussed, there are very few feasible equilibrium solutions within the statistical universe.
We can convert this simple C-R example problem into an inverse problem for the equilibrium in terms of the interactions, , by setting Eq. 2 to and rephrasing Eq. 2 in terms of the interactions not the equilibrium densities. Doing so yields:
| [3] |
with E (signifying equilibrium) the matrix specifying the equilibrium abundances, which leaves us with an underdetermined system of two equations with three unknowns. In this relatively simple case, after setting , treating the coefficients as the unknowns, we can solve for the exact solution set which satisfies the linear equations:
| [4] |
| [5] |
Next, we see how the underdetermined system leads (Fig. 1, 2) to the linear planar set of solutions between and (Fig. 1, 3; blue planar solutions). Immediately the rephrasing has moved us to feasible subset of the statistical universe. Now, the inverse problem, phrased such, also conveniently allows us to add additional biological constraints directly on the interaction strengths (Fig. 1, 4). It is a major energetic constraint in model systems that the negative impacts of the consumer on the resource must be larger or equal to the benefit the consumer receives from this interaction (energetically, there is imperfect conversion of resource to consumer). This fundamental energetic constraint is expressed as (if abundances are measured in terms of biomass):
| [6] |
Substituting Eqs. 4 and 5 and rearranging in terms of a11 yields:
| [7] |
With these biological constraints (feasibility and energetic), the system goes from the extremely large volume of solutions for , to the bounded line of solutions above (Fig. 1). This line represents a set of parameters given equilibrium densities that produce a feasible and energetically constrained C-R. Randomly choosing equilibrium densities thus puts us in the position to calculate parameters that generate energetically constrained feasible C-R systems. With the calculated parameters and the equilibrium densities we can then determine the community matrices and calculate stability using standard numerical algorithms (i.e., calculate maximum real part of all eigenvalues). In this case, after many random samples drawn from the constrained parameter space, we can use the stability results and the known equilibrium densities to interpret this stability result. Specifically, different random equilibrium draws create a range of C:R biomass ratios (i.e., steepness of the C-R biomass pyramid) showing us that steep or top-heavy C-R (x2/x1) biomass pyramids are inherently less stable—a result entirely consistent with much existing low-dimensional theory (25). The simple inverse rephrasing of the problem, more numerically solvable, allows a unique and very general entry point into this longstanding problem.
Scaling to High-Dimensional Food Webs.
Having shown the approach for a 2-dimensional consumer-resource model, the extension to higher dimension is straightforward conceptually but much more difficult computationally. As above, we start off with the assumption that we know all equilibrium biomass densities and then we are free to invoke constraints for all interactions in the diverse webs. Various additional biological constraints can be added (e.g., more precise energetic relationships between consumer–resource that follow some stronger assumptions). Our methodology, as discussed above, harnesses a recent bioinformatic approach (see methods for details) but otherwise is identical to the consumer–resource ideas in the simplest model, as encapsulated in Eqs. 1–7.
Results
With the bioinformatic inverse approach in hand, we now can answer the question of how large, diverse, stable, food webs can arise. We do this by considering the three following informative cases highlighted in the “A Concrete Example” section above: i) the unconstrained statistical universe model (e.g., Fig. 2A); the feasible equilibrium constraint (Fig. 2B), and the feasible constraint plus the energetic constraint (Fig. 2C). This combination allows us to look at the changing stability response to diversity (Fig. 2) over the gradient in increasing biological constraints while also harnessing the inverse methodology to examine the “filtered” interaction strength structures left behind (for both diagonal self-limitation and off-diagonal distributions; Fig. 3). We will end by then showing that the most biological constrained case shows filtered interaction strengths distributions that generate a generalist–specialist interaction strength trade-off that likely mediates stable feasible high-diversity webs (Fig. 4)—a trade-off that has been repeatedly posited empirically (33).
Fig. 2.
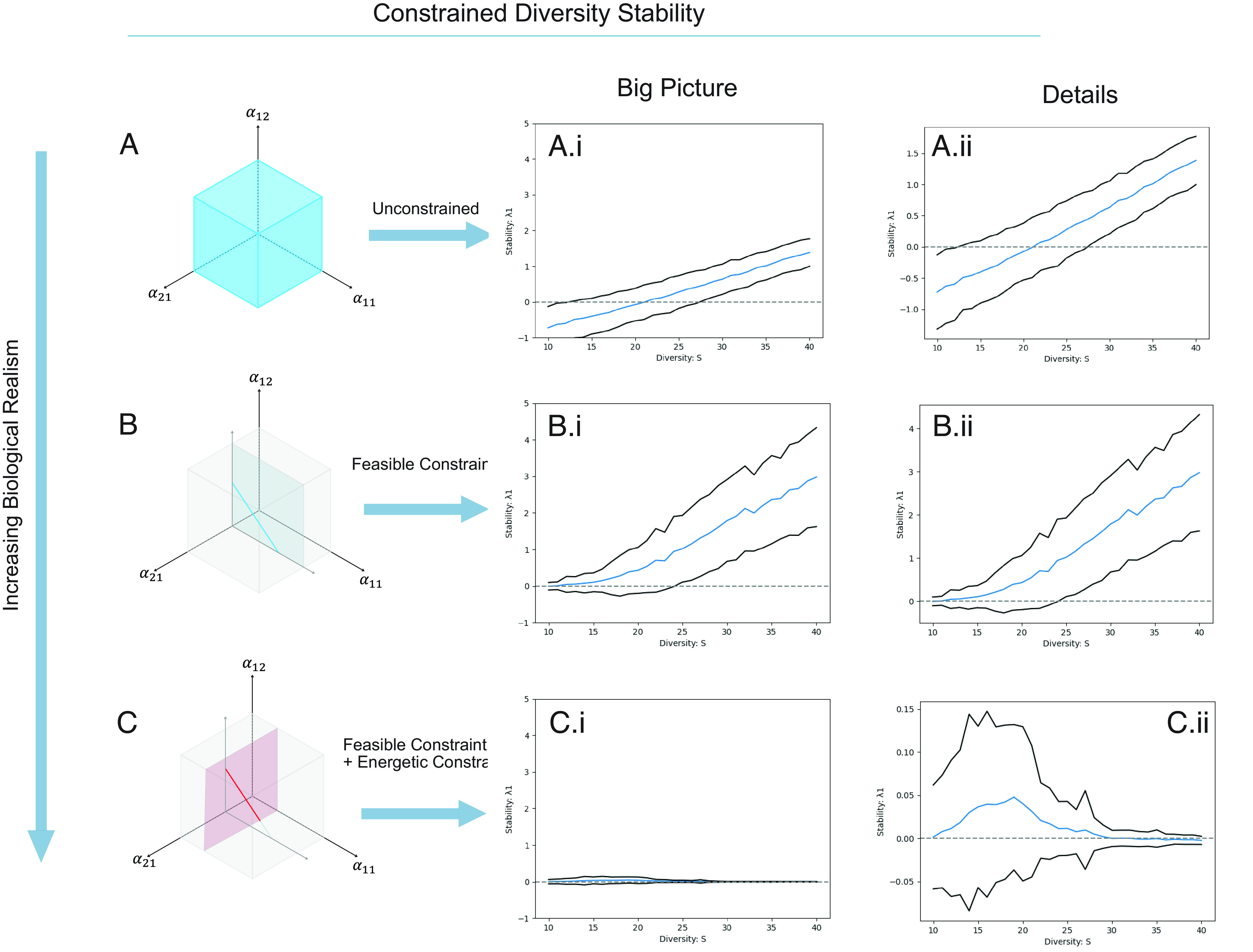
Stability responses to diversity across a range of biologically constrained whole food web models. These changes in constraint level effectively consider all of the parameter space (here represented as an example cube; see A Concrete Example C-R section in the text), a feasibility slice (lower dimensional slice of parameter space represented as a plane) and even lower dimensional slice through the parameter space (here represented as a line). The mean stability response [solid curves, Re(λMAX)] and the 95% CI stability to diversity of (A) unconstrained random matrix models; (B) inverse method matrix model under only feasibility constraint, and; (C) inverse method matrix model under feasibility constraint and energetic constraint (i.e., imperfect conversion). Panel (i) shows the same scaling of the stability responses while panel (ii) zooms in to show the more detailed response of each case. Classical, unconstrained models (A, i, and ii) and feasibility alone models (B, i, and ii) show the characteristic destabilization curves with diversity while the energetically constrained model shows a decoupled stability response with diversity and a relatively consistent proportion of stable webs. We note in this case they are relatively weakly stable or relatively weakly unstable and do not experience the extreme instability of the classical and feasible model.
Fig. 3.
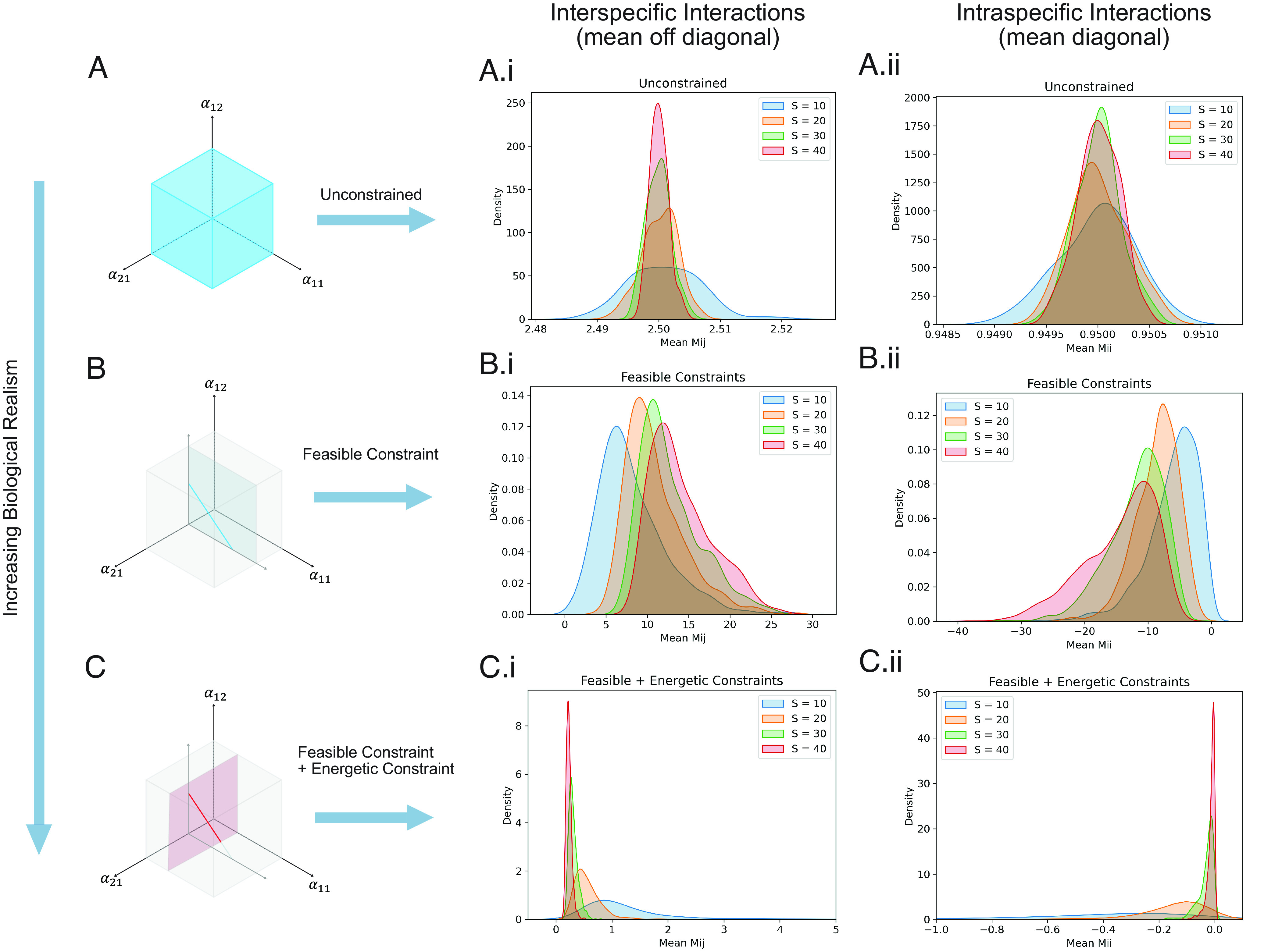
Distributional responses of interaction strength to diversity across a range of biologically constrained whole food web models for the unconstrained (A); the feasibility constrained (B) and the feasibility + energetically-constrained case (C). The distributional responses of the interaction strengths (green to blue) display the off-diagonal interspecific distributions [i.e., column marked (i)] and the off-diagonal intraspecific distributions [column marked (ii)]. The unconstrained random matrix models do not shift with diversity as expected (A, i and ii); the inverse method matrix models under only feasibility constraint shows a shift with increasing diversity to stronger interactions (B, i, and ii), and; inverse method matrix model under feasibility constraint and energetic constraint (i.e., imperfect conversion) show a strong shift towards weaker interactions. Note, the distributions occur for all webs (stable and unstable) under the different cases (unconstrained, feasible, feasible, and energetic). (C, i, and ii). Here, the energetically constrained case is effectively operating to select weak regulation. Note, the distributional shifts of C, i, and ii for the highly constrained case suggest that webs do not require increased self-regulation to be stable and here energetic constraints are associated with feasible solution that rely mostly on weak interactions. All distribution plots use automatic kernel smoothing with a Gaussian kernel. Manual spot checks were used to see if nearby kernels have significantly different shapes. We detected no such dependence.
Fig. 4.
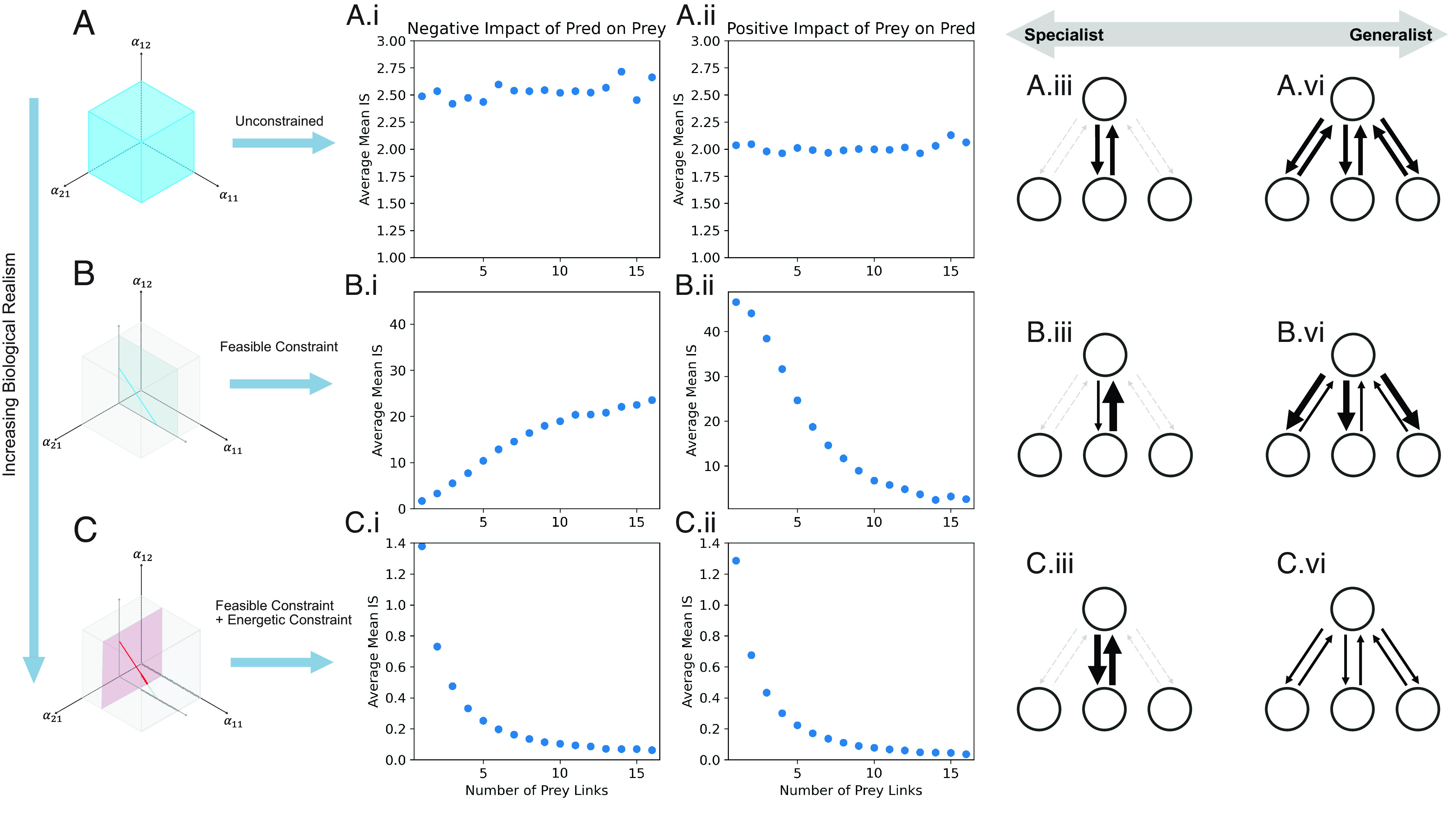
Interaction strength relationship with generality of feeding. Here, we look at the average interaction strength for species with different numbers of feeding links (i.e., different degrees of generality) for (A) Unconstrained; (B) Feasible Constraints; (C) Feasible + Energetic Constraints all for food webs of 40 species. Models C, energetically constrained webs, alone shows reduction in mean IS with increasing generality. Intriguingly purely feasible constraints (B) show opposite effects of increasing generalism, leading to ever increasing negative impacts as prey links increase, while the benefit being conferred to the predator being reduced.
Our first result is that the model with the most biological plausible constraints (i.e., both feasible and energetically constrained Fig. 2C) is the only model capable of consistently producing a high fraction of stable webs (Fig. 2 A–C). Interestingly, the addition of the energetic constraint appears to strongly filter out both highly stable and highly unstable webs. Fig. 2 A–C, i show these results for the same local stability range, while Fig. 2 A–C, ii show the same results but with zoomed in local stability scales. Indeed, the most constrained webs show a strong tendency to hover near local stability at all times (i.e., weakly stable or weakly unstable; Fig. 2 C, i and ii), precluding the well-known destabilizing outcome of increasing diversity that the unconstrained (Fig. 2 A, i and ii) or less constrained (feasible) webs produce (Fig. 2 B, i and ii), Thus, our most biologically constrained web yields a consistently stable or nearly stable, set of solutions for more diverse webs (i.e., >20 species Fig. 2 C, ii), in stark contrast to the classical unconstrained (Fig. 2 A, ii) and feasible (Fig. 2 B, ii) model, which both become highly unstable [two orders of magnitude change in dominant Re(λ)] for the same network topologies (P) and distributions of biomass (x) and growth rate (r) (Fig. 2 A and B).
To understand how the addition of biological constraints filter the interaction strength distributions of the food web models, we examined the mean and variance of the distribution for the above diversity–stability experiments. In Fig. 3, we display a column with similar y-axis scaling (mean interaction strength or variance in interactions) and a zoomed in y-axis scaling to allow the details of different biological filters to emerge. Our second set of major results is that the biological filters of the inverse method act to strongly modify the off-diagonal interaction strengths (i.e., interspecific interactions) and the diagonal interaction strengths (intraspecific interactions) compared to the classical matrix results (Fig. 3 A–C).
To see this, note that the classical matrix results effectively show unfiltered interaction strengths as the distributions for each new diversity sampling literally lie on top of each other for i) the mean interspecific interaction strengths (Fig. 3 A, i; unconstrained), and; ii) the mean diagonal or intraspecific interaction strengths (Fig. 3 A, ii; unconstrained). On the other hand, the feasibility constraint, and the energetic constraints (recall also only feasible solutions) show strong filtering of structure with diversity (Fig. 3 B and C). Curiously, the “feasible only” case shows that interspecific interactions strength increases with diversity (Fig. 3 B, i), but this occurs with a strong increasingly negative shift in the diagonal intraspecific interaction strengths (Fig. 3 B, ii). In effect, diverse feasible webs are strongly muted by the diagonals as argued in classical diversity–stability theory (34). Finally, the addition of the energetic constraint, or highly constrained model, yields quite different food web structural results. Recalling from Fig. 2, this most constrained case also has a relatively high proportion of stable, diverse webs (Fig. 2 C, i and ii). In this case, interspecific interaction strengths weaken significantly with diversity (Fig. 3 C, i) as suggested in the literature (10, 14, 35), while the diagonal terms become increasingly less negative (i.e., less damped; Fig. 3 C, ii). Note here, that theory suggests that the stabilizing role of the growing number of weak interactions must therefore overwhelm the well-known destabilizing effect of reduced diagonal damping. To more fully check this, we examined the properties of the Gershgorin circles most likely associated with the maximum real eigenvalue (and thus the stability result; see SI Appendix, Diagonal and Off diagonal Impacts S1). Here, as diversity increases, the diagonal element associated with the maximum Gershgorin circle (i.e., the circle with the most positive extent) in fact gets smaller suggesting that it is unlikely that a single strong negative diagonal is mediating this result. The weakening of self-regulation with increasing diversity in this stability result is noteworthy and may be why the local stability in the most constrained model is ultimately relatively weakly stable. In both constrained models, the variance in diagonals follow the same pattern as the variance in the mean of the interspecific interactions.
To further explore what is potentially yielding the relatively high proportion of stable diverse solutions in our most biologically constrained models (energetic versus classical; Figs. 2 and 3), we looked to see if the food web structures intrinsically produced trade-offs found in real ecosystems. Since interactions were shown to weaken with diversity in this case—as argued both by low-diversity modular theory and high-diversity theory (14, 35)—we looked to see if a generalist (weak)–specialist (strong) interaction strength trade-off manifested. As expected, random models showed no patterning between the number of prey links and mean strength (for positive or negative impact of predator on prey interaction strengths) as shown here for the high–diversity case (Fig. 4A). On the other hand, feasibility-constrained models alone produced biologically odd solutions. Specialists (Fig. 4B) shows strong positive interaction strengths of prey on predator but extremely muted negative effects on their prey (energetically impossible). As the number of links grow, the more generalist species have massively strong average interaction strengths for predator on prey (negative) but extremely muted positive effects of prey on predator (Fig. 4B). This may allow feasibility but it is entirely inconsistent with known biological constraints. Finally, and consistent with our empirical understanding, the energetically constrained models produce strong specialist–generalist trade-offs such that specialists impart strong positive and negative interaction strengths and generalists are governed by more muted interaction strengths (Fig. 4C). Thus, our final result is that the energetically constrained model produces configurations that tend to show dramatic weaker interaction sets for more generalized species and stronger interactions in species that are more specialized (Fig. 4). This trade-off has long been argued to be a potent stabilizing mechanism in food webs, readily producing weak interactions capable of muting destabilizing strong interactions and here the inverse methods with constraints effectively demonstrate this trade-off.
Discussion
The question of the existence of large complex ecosystems has puzzled ecologists for a long time since classical work that suggested that the chance of finding a stable complex food web by random search among all possible webs of interaction was small (8). Here, following other applications of the inverse method in ecology (22), we have turned the problem on its head (i.e., an inverse problem), and started instead with the large complex web as a given and asked what properties (meaning interactions among species) it must have. We then employed numerical approaches identical to recent bioinformatic advances in metabolic network research that had faced an analogous inverse computational problem (23).
This network approach has two major theoretical advantages over the classical approach started by Robert May over 40 y ago. First, the approach only produces results for feasible webs, which correspond to the species all being at a positive equilibrium, a problem the classical matrix approach frequently ignores [although see (14, 16)]. Second, the approach allows one to posit biological structure that operates to maintain the local stability of diverse real webs. Not only does this approach have these potential advantages, it produces new insights into the maintenance of complex, diverse food webs. With this tool in hand, we compare classical random matrices with realistic topology to increasingly constrained matrices (i.e., feasible and feasible-energetically constrained matrices). We find that the most biologically constrained models yield a consistently larger fraction of stable high-diversity webs in contrast to the presence of highly unstable diverse webs known to occur from the classical random matrix approach with known topological structure. More specifically, feasible energetically constrained (i.e., requiring imperfect energy conversion) webs tend to produce more stable high-diversity webs than the random webs known to be composed mostly of infeasible matrices.
While Robert May’s approach operated with a statistical universe (all interaction strengths within some bounded set, see Fig. 2A), the inverse approach has the novel property that it allows us to only look at the collection of webs corresponding to realistic feasible solutions. Here, we use this intriguing filtering property to show that increasing biological constraints suggest that more diverse webs do not require strong self-regulation as the classical methods seem to require and that these constrained webs show that an increasing preponderance of weak interactions accompany more diverse feasible webs (Fig. 3). We note, though, that consistent with the literature that argues consistent regulation is required for stability (33), these same diverse webs all show consistent evidence of weak regulation by all species. Finally, we examine the feasible webs to show that they naturally create biologically constrained matrices that produce a generalist–specialist trade-off whereby generalist predators have weaker interactions than more specialized species.
Akin to the classical matrix method that has been successfully producing theory for over 50 y, the inverse approach is rich in potential for theoretical advances. Here, we have used this approach to rephrase the longstanding diversity–stability problem for ecological food webs and in doing so revealing patterns in interaction strength distributions that bely the stability responses. Clearly, we have only scratched the surface of realistic and interesting biological constraints exist. Recent developments, for example, suggest that the shape of the Eltonian pyramid (or lack thereof) drive stability and the inverse methodology with known equilibrium densities is well positioned to investigate this (i.e., bottom heavy to top heavy as per the C-R example). Additionally, the ability to input real data in the form of biomass equilibrium estimates, with additional plausible biological constraints, allows an empirical entry point that interfaces with diversity–stability theory—an area of high-diversity food webs that has always been lacking.
Materials and Methods
To conduct simulations of constrained random and biological food webs, we implemented a Hit and Run sampler in the Julia programming language (36) following the artificial centering algorithm described in ref. 37 based on the implementation found in the Python CORBA toolkit (38), as well as using the Gurobi optimization platform (39) so that the algorithms can be run in parallel (for each independent Hit and Run MC chain). This is a computationally intensive procedure so high-performance algorithms and languages are required. The models ran on a 12-core machine so 12 Hit and Run Monte Carlo sampling chains were generated and run in parallel at a given time to achieve the desired output.
Generation of Constrained Food Webs.
The core method requires building the Inverse Problem from the input of three pieces of biological information: 1) the predation network, P, a matrix of (0, 1) describing which species feed on which other species, this is generated from an ecological network model, the Generalized Cascade Model (40). This model has been found to recreate common empirical findings for network structure across diverse ecosystems but does not introduce compartmentation like the more popular Niche model (41, 42) which we wanted to control for to make the results more comparable to classical random matrix approaches, while still being biologically meaningful. 2) A biomass value for each species in the food web, x, we outline the procedure we employed in the sampling methods below. 3) For each species in the food web either the instantaneous growth rate (+ value), if the species is basal, or the per capita mortality if the species is nonbasal (− value). This follows the basic feasibility assumption as outlined by (14) for basal versus predator species in random matrix models (or any model based on Generalized Lotka-Volterra equations). We collect this information into the vector, r.
With these three pieces of information, we can then produce the Inverse Problem . Here, E is determined by the predation matrix, P, with the convention that the unknown, nonzero, pairwise interaction strength parameters , are built by row-wise concatenation (i.e., ), where S is the number of species in the food web. To maintain the equality relationship of the Inverse Problem, entries in E, are made by forming a block diagonal matrix with the diagonal blocks being the associated biomass entries such that for the nonzero elements of the nth row of , we take the associated nonzero indices (js) and take the element into E. For example, given the matrix:
and biomass vector , first we produce the vector by concatenating the nonzero entries of A row wise. Then, row 1 of has nonzero elements the corresponding column indices (1, 2); therefore, we have first block of E equal to . Similarly for the second row, we have the nonzero elements being with correspoinding column indices (1, 2, 3), giving the second block of equal to , finally the only nonzero element in the third row of is with column index (2), given their block of as , yielding the block diagonal matrix:
This inverse problem determines the equality constraints required for the network structure, biomass, and growth/morality rates to be feasible. We further allow inequality constraints to be defined on any linear combination of the vector. In our model, we have simple energetic constraints that representing the ecological constraint that the per capita growth potential a predator receives from its prey cannot be larger than its impact.
With the equality and inequality constraints, we then can use a linear programming optimization routine to determine if the constrained random food web has a nonempty solution (i.e., a nondegenerate polytope in the solution space). As we are using this approach to be in line with random matrix approaches, each of our inputs are random, though biologically motivated/constrained, we can have a situation where no solution fits the particular triple (P, x, r). In which case, we draw a new random set of equilibrium conditions and test again. We allowed for up to 1,000 restarts, and that was sufficient to always find a feasible starting configuration (i.e., we found parameters that solved for the equilibrium so we found a feasible solution).
Once we have a nonempty solution space for the Inverse Problem we proceed to sample the set of solutions using the Hit and Run algorithm.
Sampling Procedure.
The Hit and Run sampler we are using returns a vector of the nonzero elements of the matrix . This parameterization is a bit awkward to use for biological models so it is common to build numerical libraries that can move between the biological representation of the problem and the low-level form needed by the numerical methods (38). The approach used by standard libraries like COBRApy are tailored for metabolic networks, so we first built tools that transformed a Generalized Lotka Volterra (GLV) system into the form needed by the sampler, and back (from the vector our sampled parameters, back into a GLV form for analysis). The code for this is available at https://github.com/gabrielgellner/constrained-foodwebs.
Specifically, we start with a system in the generalized Lotka-Volterra form , with being the species diversity of the system, , the matrix of per capita interaction terms, , the length vector of equilibrium biomass/density for each species, and is the length vector of growth and death rates (43). The algorithm requires inputs for the and vectors, and the Hit and Run sampler will return a uniform sample from (32, 37).
To generate the biomass vector, we first use the random predation matrix, , to calculate the binary trophic position using Levine’s algorithm (44) and save this information for each species in the array . Then biomass pyramid parameter, , is randomized between −10 and 10 to model top-heavy webs ( ) and bottom-heavy ( ) biomass equilibrium instead of just have biomass structures with not pyramid structure, like a purely uniform distribution would. Specifically, we have the specific biomass determined as if and if .
For the vector, we first determine which species are basal using the TP calculation from above and then set all basal species to have intrinsic growth rates of and all nonbasal species to have mortalities of .
We ran each sampler chain for 10^6 iterations, saving only the last 5 × 10^5 samples to allow burn in. We then shuffled the resulting samples to remove serial correlation as described in ref 37. We generated 5 × 10^5 samples 1,200 times per species diversity level, S, ranging from 10 to 40.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Funding by “Food from Thought” (499075) to G.G. and K.M., NSERC Discovery to K.M. (400353) and NSF Grant DMS-1817124 to (A.H.).
Author contributions
G.G., K.M., and A.H. designed research; G.G., K.M., and A.H. performed research; G.G. contributed new reagents/analytic tools; and G.G., K.M., and A.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: B.B., Carl von Ossietzky Universitat Oldenburg Institut fur Chemie und Biologie des Meeres; and M.N., Oregon State University.
Data, Materials, and Software Availability
There are no data underlying this work.
Supporting Information
References
- 1.Polis G., Complex trophic interactions in deserts: An empirical critique of food-web theory. Am. Nat. 138, 123–155 (1991). [Google Scholar]
- 2.Sax D. F., Gaines S. D., Species diversity: From global decreases to local increases. Trends Ecol. Evol. 18, 561–566 (2003). [Google Scholar]
- 3.Blanchard J. L., A rewired foodweb. Nature 527, 173–174 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bartley T. J., et al. , Food web rewiring in a changing world. Nat. Ecol. Evol. 3, 345–354 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kortsch S., Primicerio R., Fossheim M., Aschan D. A. V. M., Climate change alters the structure of arctic marine food webs due to poleward shifts of boreal generalists. Proc. R. Soc. B 282, 20151546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuelson P., Foundations of Economic Analysis (MIT Press, 1947). [Google Scholar]
- 7.Gardner M. R., Ashby W. R., Connectance of large dynamic (cybernetic) systems: Critical values for stability. Nature 228, 784 (1970). [DOI] [PubMed] [Google Scholar]
- 8.May R. M., Will a large complex system be stable? Nature 238, 413–414 (1972). [DOI] [PubMed] [Google Scholar]
- 9.McNaughton S. J., Diversity and stability of ecological communities: A comment on the role of empiricism in ecology. Am. Nat. 111, 515–525 (1977). [Google Scholar]
- 10.Yodzis P., The stability of real ecosystems. Nature 289, 674–676 (1981). [Google Scholar]
- 11.Haydon D. T., Pivotal assumptions determining the relationship between stability and complexity: An analytical synthesis of the stability-complexity debate. Am. Nat. 144, 14–29 (1994). [Google Scholar]
- 12.Allesina S., Tang S., The stability-complexity relationship at age 40: A random matrix perspective. Popul. Ecol. 57, 63–75 (2015). [Google Scholar]
- 13.James A., et al. , Constructing random matrices to represent real ecosystems. Am. Nat. 185, 680–692 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Emmerson M. C., Yearsley J. M., Weak interactions, omnivory and emergent food-web properties. Proc. Biol. Sci. 271, 397–405 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilpin M. E., Stability of feasible predator-prey systems. Nature 254, 137–139 (1975). [Google Scholar]
- 16.Grilli J., et al. , Feasibility and coexistence of large ecological communities. Nat. Commun. 8, 14389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brose U., Williams R. J., Martinez N. D., Allometric scaling enhances stability in complex food webs. Ecol. Lett. 9, 1228–36 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Iles A. C., Novak M., Complexity increases predictability in allometrically constrained food webs. Am. Nat. 188, 000–000 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Gellner G., McCann K. S., Consistent role of weak and strong interactions in high- and low-diversity trophic food webs. Nat. Commun. 7, 11180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allesina S., Tang S., Stability criteria for complex ecosystems. Nature 483, 205–208 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Christensen V., Pauly D., ECOPATH II—A software for balancing steady-state ecosystem models and calculating network characteristics. Ecol. Modell. 61, 169–185 (1992). [Google Scholar]
- 22.Van Oevelen D., et al. , Quantifying food web flows using linear inverse models. Ecosystems 13, 32–45 (2010). [Google Scholar]
- 23.Schellenberger J., Palsson B., Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 284, 5457–5461 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan S., Cluett W. R., Mahadevan R., Constructing kinetic models of metabolism at genome-scales: A review. Biotechnol. J. 10, 1345–1359 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Mccauley D. J., et al. , On the prevalence and dynamics of inverted trophic pyramids and otherwise top-heavy communities. Ecol. Lett. 21, 439–454 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Brualdi R., Shader B., Matrices of Sign-Solvable Linear Systems (Cambridge University Press, ed. 1, 2009). [Google Scholar]
- 27.Levins R., The qualitative analysis of partially specified systems. Ann. N. Y. Acad. Sci. 231, 123–138 (1974). [DOI] [PubMed] [Google Scholar]
- 28.Lawlor L. R., A comment on randomly constructed model ecosystems. Am. Nat. 112, 445–447 (1978). [Google Scholar]
- 29.May R. M., Stability and Complexity in Model Ecosystems (Princeton University Press, 1973). [PubMed] [Google Scholar]
- 30.Goh B., Jennings L., Feasibility and stability in randomly assembled lotka-volterra models. Ecol. Modell. 3, 63–71 (1977). [Google Scholar]
- 31.Lovász L., Vempala S., Hit-and-run from a corner. SIAM J. Comput. 35, 985–1005 (2006). [Google Scholar]
- 32.Megchelenbrink W., Huynen M., Marchiori E., optGpSampler: An improved tool for uniformly sampling the solution-space of genome-scale metabolic networks. PLoS One 9, e86587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson D. S., Yoshimura J., On the coexistence of specialists and generalists. Am. Nat. 144, 692–707 (1994). [Google Scholar]
- 34.Barabás G., Michalska-Smith M. j., Allesina S., Self-regulation and the stability of large ecological networks. Nat. Ecol. Evol. 1, 1870–1875 (2017). [DOI] [PubMed] [Google Scholar]
- 35.McCann K. S., Hastings A., Huxel G. R., Weak trophic interactions and the balance of nature. Nature 395, 794–798 (1998). [Google Scholar]
- 36.Bezanson J., Edelman A., Karpinski S., Shah V. B., Julia: A fresh approach to numerical computing. arXiv [Preprint] (2014), 10.48550/arXiv.1411.1607 (Accessed 6 January 2022). [DOI]
- 37.De Vericourt F., et al. , Direction choice for accelerated convergence in hit-and-run sampling. Oper. Res. 46, 84–95 (1998). [Google Scholar]
- 38.Ebrahim A., Lerman J. A., Palsson B. O., Hyduke D. R., COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst. Biol. 7, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurobi Optimization, L. L. C., Gurobi Optimizer Reference Manual (2021).
- 40.Stouffer D. B., Camacho J., Guimerà R., Ng C., Quantitative patterns in the structure of model and empirical food webs. Ecology 86, 1301–1311 (2005). [Google Scholar]
- 41.Williams R. J., Martinez N. D., Simple rules yield complex food webs. Nature 404, 180–183 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Guimerà R., et al. , Origin of compartmentalization in food webs. Ecology 91, 2941–2951 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Remien C. H., Eckwright M. J., Ridenhour B. J., Structural identifiability of the generalized lotka-volterra model for microbiome studies. R. Soc. Open Sci. 8, 201378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine S., Several measures of trophic structure applicable to complex food webs. J. Theor. Biol. 83, 195–207 (1980). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
There are no data underlying this work.


