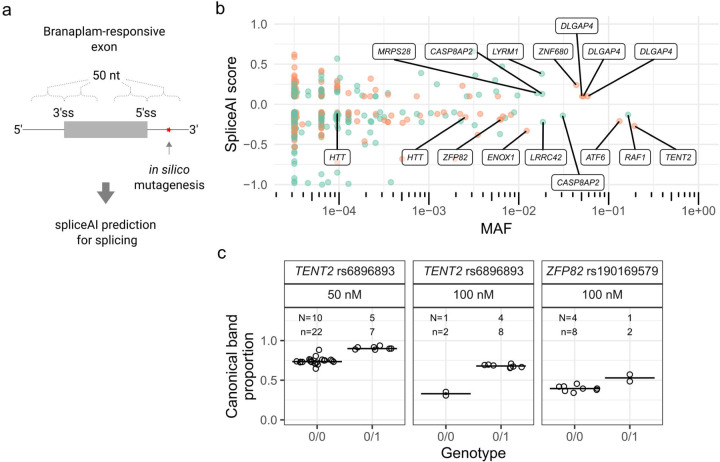
Figure 3.
SpliceAI identified variants predicted to affect splicing of genome-wide branaplam-responsive exons. (a) SpliceAI predictions were made for variants within 50 nt of branaplam-responsive exon and pseudoexon splice junctions. (b) Variants near branaplam-responsive pseudoexons (orange) and exons (green) that yield significant SpliceAI scores are plotted by allele frequency with gene names indicated for selected variants. HTT variants rs148430407 (MAF 2.6x10−3) and rs772437678 (MAF 9.6x10−5) are labelled, while rs145498084 did not have a significant SpliceAI score (c) SpliceAI-predicted variants affect splice modulation of TENT2 and ZFP82. Proportion of canonically spliced product across tested LCLs for TENT2 and ZFP82, grouped by no presence (0/0) or heterozygous presence (0/1) of variant. N = Number of cell lines for variant, n = cultures analyzed.