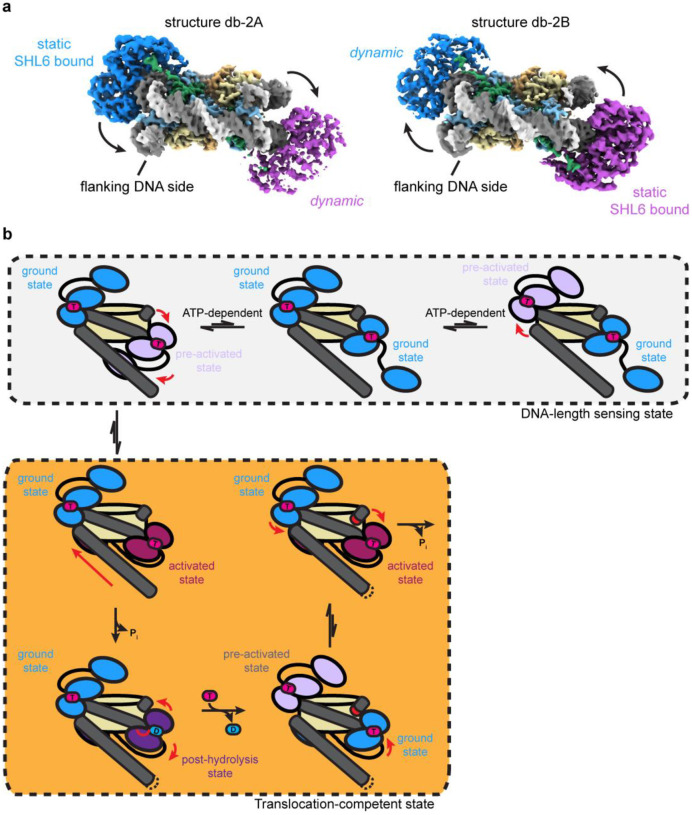
Fig. 4. Coordinated asymmetric actions of SNF2h protomers on a nucleosome.
a) Two coulomb potential maps representing the endpoints of one principal component from 3DVA of the double-bound SNF2h-nucleosome particles. On one end (structure db-2A), one SNF2h is stably bound to the nucleosome, while the other SNF2h is more dynamic. On the other end (structure db-2B), the SNF2h that was stably bound to the nucleosome becomes more dynamic, while the SNF2h that was more dynamic becomes more static.
b) Working model for SNF2h conformations while coordinating actions on a nucleosome. In the DNA-length sensing state, each SNF2h protomer can either be in a ground state or dynamic pre-activated state while searching for flanking DNA using its HAND-SANT-SLIDE (HSS) domain. The protomer that is able to sense flanking DNA will undergo further conformational change to reach an activated state that promotes ATP hydrolysis and DNA translocation. Exchange of ADP for ATP resets SNF2h to a ground state, and the other SNF2h protomer then has first priority to again search for flanking DNA in a dynamic, pre-activated state.