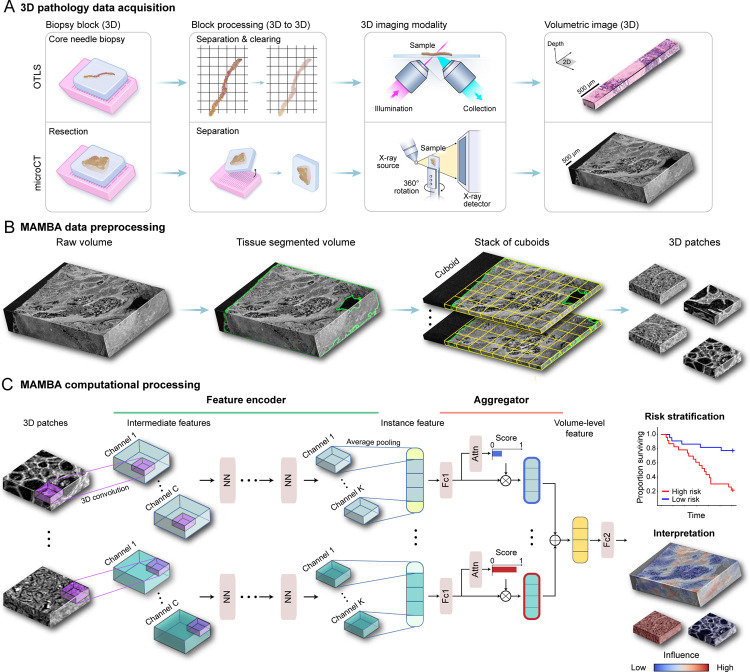
Figure 1: MAMBA computational workflow.
(A) With 3D imaging modalities such as open-top light-sheet microscopy (OTLS) and microcomputed tomography (microCT), high-resolution volumetric images of tissue specimens are captured. (B) MAMBA accepts raw volumetric tissue images from diverse imaging modalities as inputs. MAMBA first segments the volumetric image to separate tissue from the background. In a common version of the workflow, the segmented volume is then treated as a stack of cuboids (3D planes) and further tessellated into smaller 3D patches. (C) The patches (i.e., instances) are then processed with a pretrained feature encoder network of choice, leveraging transfer learning to produce a set of compact and representative features. The encoded features are further compressed with a domain-adapted shallow, fully-connected network. Next, an aggregator module aggregates the set of instance features, automatically weighing them according to the importance towards rendering the prediction to form a volume-level feature. MAMBA also provides saliency heatmaps for clinical interpretation and validation. The computational workflow of MAMBA with 2D processing is identical. Further details of the model architecture are described in the Methods. NN, generic neural network layers dependent on the choice of feature encoder; Channel C, K, intermediate channels in feature encoder; Attn, attention module; Fc1, Fc2, fully-connected layers.