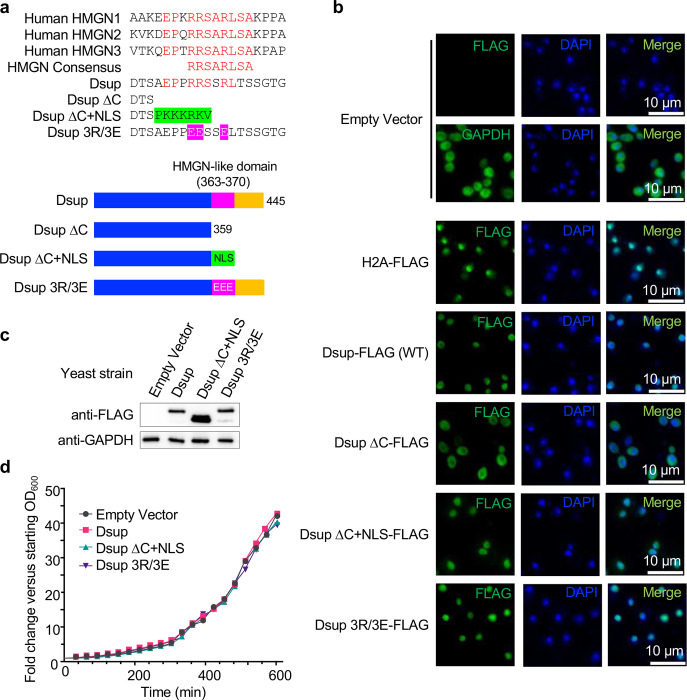
Fig. 2. Heterologous Dsup is nuclear localized in yeast and does not negatively impact growth rate.
(a). Alignment of the Dsup HMGN-like domain (aa 363–370, RRSSRLTS), mutants of same from this study, and the HMGN core consensus (RRSARLSA) derived from human HMGN1–3. Residues in red are identical between human HMGN1–3, the HMGN core consensus and Dsup. Green indicates the nuclear localization signal (NLS: PKKKRKVPKKKRKV) added onto the Dsup ΔC construct. Pink indicates three arginine to glutamic acid substitutions (R363E/R364E/R367E) in the HMGN-like sequence to create Dsup 3R/3E (Figure adapted from17). Beneath are schematics of the Dsup wild-type and mutant alleles (all containing N-terminal 6xHIS and C-terminal FLAG tags; not depicted). (b). Immunofluorescence to examine the subcellular location of Dsup alleles (anti-FLAG) in yeast. DAPI co-staining identifies nuclei. H2A-FLAG and GAPDH are respective controls for nuclear and cytoplasmic localization. (c). Western blot showing relative expression of indicated Dsup alleles (anti-FLAG) in yeast. Anti-GAPDH is a loading control for each strain protein extract. (d). Representative growth curves of yeast expressing Dsup alleles.