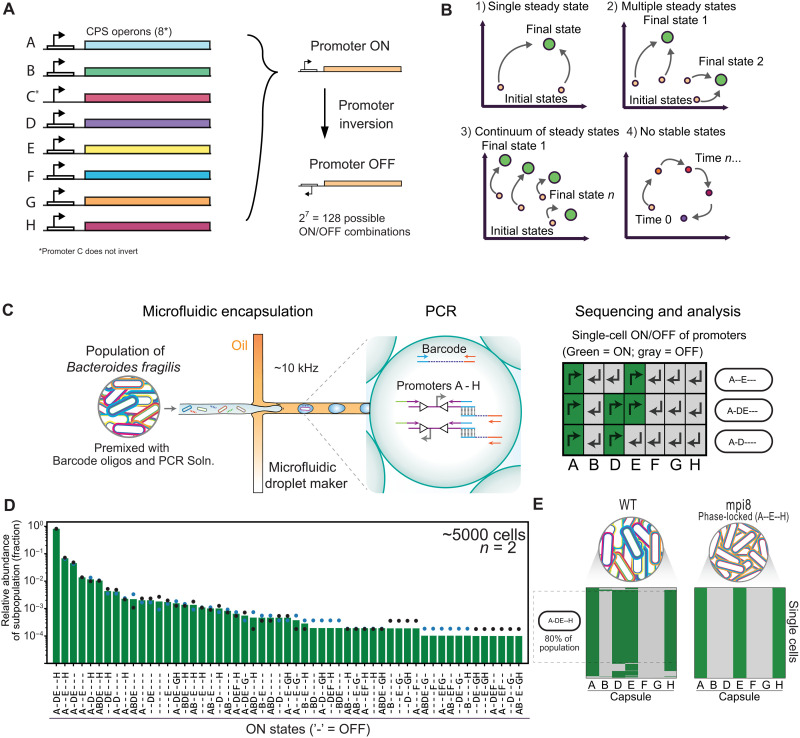
Fig. 1. High-throughput single-cell sequencing of CPS promoters in B. fragilis reveals subpopulations defined by combinatorial promoter orientations.
(A) Summary schematic of B. fragilis CPS loci. (B) Cartoon schematic of potential population composition landscapes under continuous growth. Populations could (1) converge to a single stable state, (2) converge to multiple stable states, (3) exist in a continuum of stable states, or (4) remain in flux with no stable states. (C) Cartoon representation of the ultrahigh-throughput single-cell targeted sequencing workflow for B. fragilis CPS promoters. We first premix cells from a colony or liquid culture with barcode oligos, high-fidelity PCR mix, and target locus primers (14 primers for the seven invertible CPS promoters) containing adaptor sequences that overlap the barcode oligo. Cells and barcodes in this mix are then encapsulated into picolitre emulsion droplets at limiting dilution so that most droplets contain 1 or 0 cell/barcode according to a Poisson distribution. Droplets that contain both a barcode oligo and a cell are selectively amplified and linked to genomic loci via overlap extension PCR (see fig. S1 for details). The cell is lysed during the initial PCR denaturing step. Sequencing reveals the promoter orientations of all targeted loci for thousands of cells. Right: Promoter orientation heatmap for three representative cells. The “ON” orientation for a particular CPS promoter is green. Dashes (“-”) represent promoters that are oriented OFF. (D) The CPS promoter states of a single colony of B. fragilis, shown as a bar plot. Data points represent two technical replicates. (E) Heatmaps as described in (C) for thousands of cells, with groups of rows clustered to reflect subpopulations with alternative combinatorial promoter orientations. A control strain with the primary recombinase gene (mpi) deleted displays a population with 100% of cells having the expected promoter configuration (A--E--H) for this particular isolate. WT, wild-type.