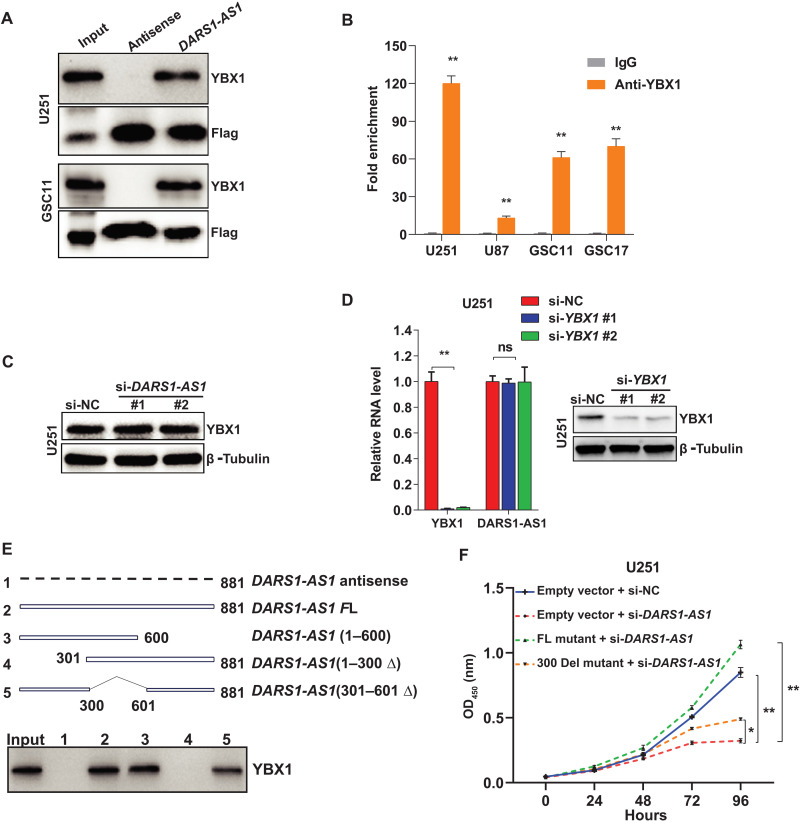
Fig. 5. Validation and characterization of the interaction between DARS1-AS1 and YBX1.
(A) RNA pull-down coupled with Western blot validated the interaction between DARS1-AS1 and YBX1 that was identified from MS analysis. (B) RIP with an anti-YBX1/anti-IgG antibody followed by RT-qPCR validated the association of YBX1 with DARS1-AS1, where anti-IgG antibody was used as a negative control. (C) The protein level of YBX1 was determined by Western blot in U251 cells transfected with the negative control siRNA (si-NC) or individual DARS1-AS1–targeting siRNAs, where β-tubulin was used as a loading control. (D) RT-qPCR analysis of YBX1 and DARS1-AS1 RNA level in U251 cells transfected with the negative control siRNA (si-NC) or individual YBX1-targeting siRNAs. Right: The YBX1 protein expression was determined by Western blot. (E) RNA pull-down of the MS2bs-tagged antisense, full-length, and serial deletion mutants of DARS1-AS1 RNA followed by anti-YBX1 Western blotting. The three serial deletion mutants of DARS1-AS1 RNA were generated by deleting 601 to 881, 1 to 300, or 301 to 600 bp, respectively. (F) U251 cells stably transduced with the vectors expressing full-length mutant DARS1-AS1 resistant to siRNAs (FL mutant), the deletion mutant with a deletion of 1 to 300 bp (300 del) or the empty vector control, were transfected with the negative control siRNA (si-NC) or siRNAs targeting DARS1-AS1 and were cultured for 4 days. The cell growth was monitored each day with CCK-8 assay. Data in (B) are shown as mean ± SD (n = 3). **P < 0.01 by Student’s t test. Data in (D) and (F) are shown as mean± SD (n = 3). **P < 0.01, *P < 0.05, or ns, not significant (P > 0.05) by one-way ANOVA with Dunnett’s multiple comparison test.