Figure 5.
DA1 lPNs encode male angular position
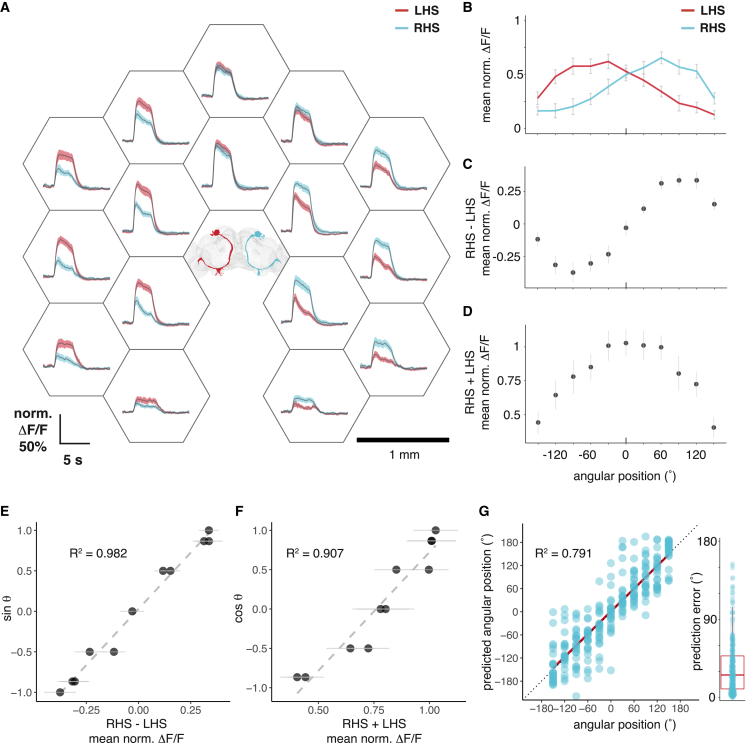
(A) GCaMP6f responses of left hemisphere (LHS, red) and right hemisphere (RHS, cyan) lPN dendrites to male presentations. Positions of the male during the stimulation protocol are indicated by the center points of the hexagons around the respective responses. The position of the imaged female is indicated by the brain (top view, facing 0°, left: negative angles, right: positive angles), and lPN colors correspond to GCaMP traces from the respective side. Average response from 8 flies, 3 trials, and shaded area is SEM of biological replicates. Scale bars, 1 mm.
(B) Angular tuning curves of left (red) and right (cyan) lPNs based on (A). Six of these positions are direct measurements, five of these (at angles 0°, ±60°, and ±120°) are based on linear interpolation at these angular directions (see STAR Methods). Error bars are SEM of biological replicates.
(C and D) The difference (C) and sum (D) of right and left mean lPN responses at given male angular positions. Error bars are SEM of biological replicates.
(E and F) The difference of right and left lPN responses correlates with the sine (E, R2 = 0.982), while the sum correlates with the cosine (F, R2 = 0.907) of the male angular position. Dashed line: linear fit.
(G) A linear model predicts male angular position based on bilateral lPN responses. Model formula in Wilkinson notation: (x, y) ∼ (lPNR − lPNL) + (lPNR + lPNL). Where x and y are the Cartesian coordinates of the male fly, and lPNR and lPNL are right and left lPN responses, respectively. The predicted angular position is calculated from the x and y predictions for single trials from (A). Predicted male angular positions correlate with the actual angular position, R2 = 0.792. Red line: linear fit, dotted line: x = y. Right: distribution of prediction error in angles, mean = 35°, median = 26°.
See also Figure S5.