Figure S7.
Integrating cVA and taste is key to controlling female receptivity, related to Figure 7
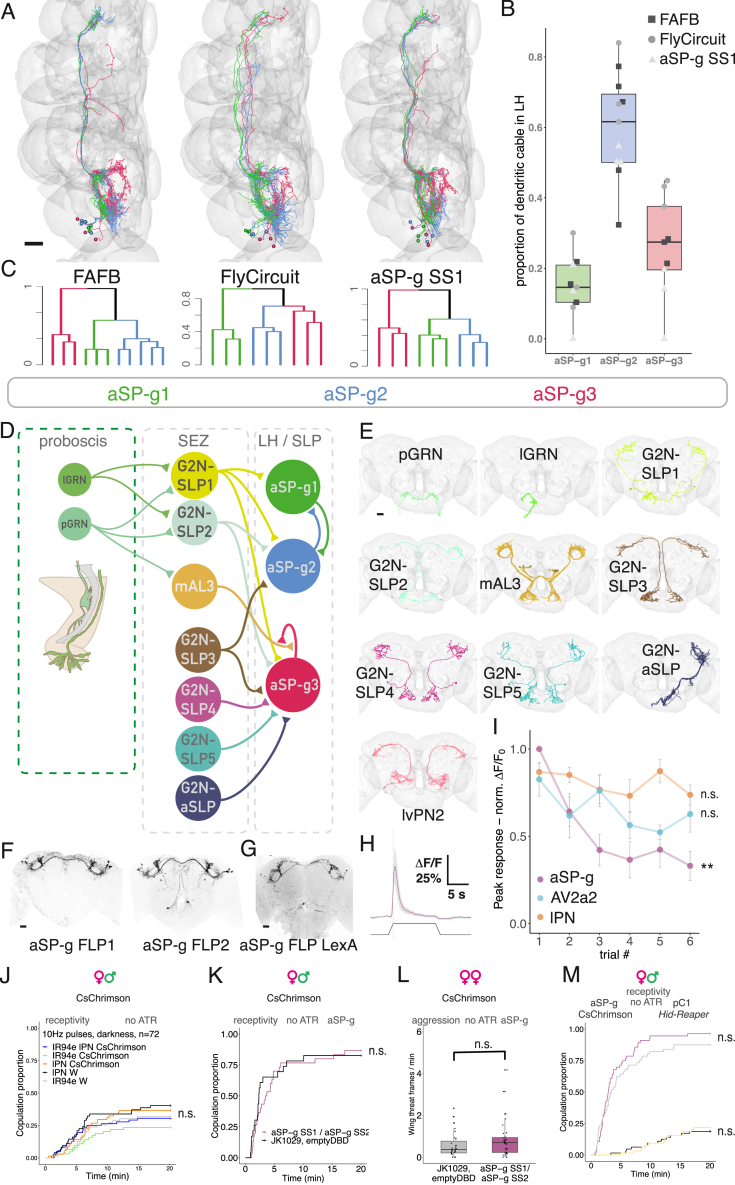
(A) Reconstructions of aSP-g neurons colored based on NBLAST morphological clustering from the FAFB dataset (left), the FlyCircuit dataset (middle), and MCFO data from aSP-g-SS (right). Clusters were named aSP-g1, aSP-g2, and aSP-g3, based on dendritic arbor position from anterior to posterior. Scale bars, 20 μm.
(B) The proportion of dendritic cable inside the lateral horn for aSP-g subtypes across three datasets. Squares: FAFB, circles: FlyCircuit, triangles: aSP-g-SS MCFO.
(C) Hierarchical clustering based on NBLAST morphological similarity scores using Ward’s method, k = 3. Order and colors are the same as in (A).
(D) Connectivity diagram of aSP-g subtypes with gustatory pathways, based on presynaptic sampling of aSP-g dendrites in the FAFB dataset, and reconstruction of G2N-SLP1 inputs via FlyWire. G2N, gustatory second-order neuron; SLP, superior lateral protocerebrum.
(E) EM reconstruction of aSP-g input neurons in the FAFB dataset. First row left, pharyngeal GRNs (pGRNs); first row middle, labellar GRNs (lGRNs); first row right, G2N-SLP1; second row left, G2N-SLP2; second row middle, mAL3; second row right, G2N-SLP3; third row left, G2N-SLP4; third row middle, G2N-SLP5; third row right, G2N-ascending SLP; fourth row, lvPN2 (cell types M_lvPNm42 and M_lvPNm44 in the hemibrain dataset); in FAFB, lPN, and lvPN2 provide 6.4% and 5.6% of aSP-g2 inputs, respectively. See Tables S3A and S3B for connectomic identifiers and synaptic weights for all other connections. Scale bars, 20 μm.
(F) Confocal image of aSP-g-FLP1 and aSP-g-FLP2 lines in a female brain, (see Table S1 for genotypes), used to block aSP-g neurons by expressing Kir2.1 (Figure 7H) maximum projection. Scale bars, 20 μm.
(G) Confocal image of aSP-g-FLP LexA in a female brain (see Table S1 for genotypes), used to activate aSP-g while ablating pC1 for neuronal epistasis (Figure 7K), maximum projection. It is important to note that aSP-g FLP-LexA weakly labels a few neurons in the midline of the brain, projecting from the peri-esophageal region to the pars intercerebralis (although the labeling of these neurons is not strong enough to appear on a maximum projection of the full brain). We cannot exclude the possibility that these neurons contribute to the behavioral effects observed in the behavioral epistasis experiment in Figure 7K. Scale bars, 20 μm.
(H) GCaMP6f responses in aSP-g axons to presenting a male at 0.75 mm distance from the antennae, female fly imaged. Mean trace from 6 flies, 6 male presentations each, gray area is SEM. Same stimulus as in Figure 2G.
(I) aSP-g responses adapt over time, unlike lPN and AV2a2. Normalized peak GCaMP6f responses to a single male presentation (as described in Figure 2G) in 6 consecutive trials separated by 35 s in three cell types, aSP-g: purple, lPN: orange, AV2a2: cyan. Responses are significantly different in aSP-g in different trials (Friedman test), but not in lPN and AV2a2. The same lPN and AV2a2 data were also used in Figures 2H, 7E, and 7F.
(J) Control for Figure 7F, using the same experimental conditions, but these flies were not raised on all-trans retinal (ATR)-containing food. n = 72 per group.
(K) Control for Figure 7G, using the same experimental conditions, but these flies were not raised on ATR-containing food. n = 32 per group.
(L) Control for Figure 7I, using the same experimental conditions, but these flies were not raised on ATR-containing food. n = 32 per group.
(M) Control for Figure 7K, using the same experimental conditions, but these flies were not raised on ATR-containing food. Notice that pC1 ablation impaired female receptivity regardless of ATR as the ablation is not temporally controlled. n = 64.
Throughout the figure, n.s. p > 0.05.