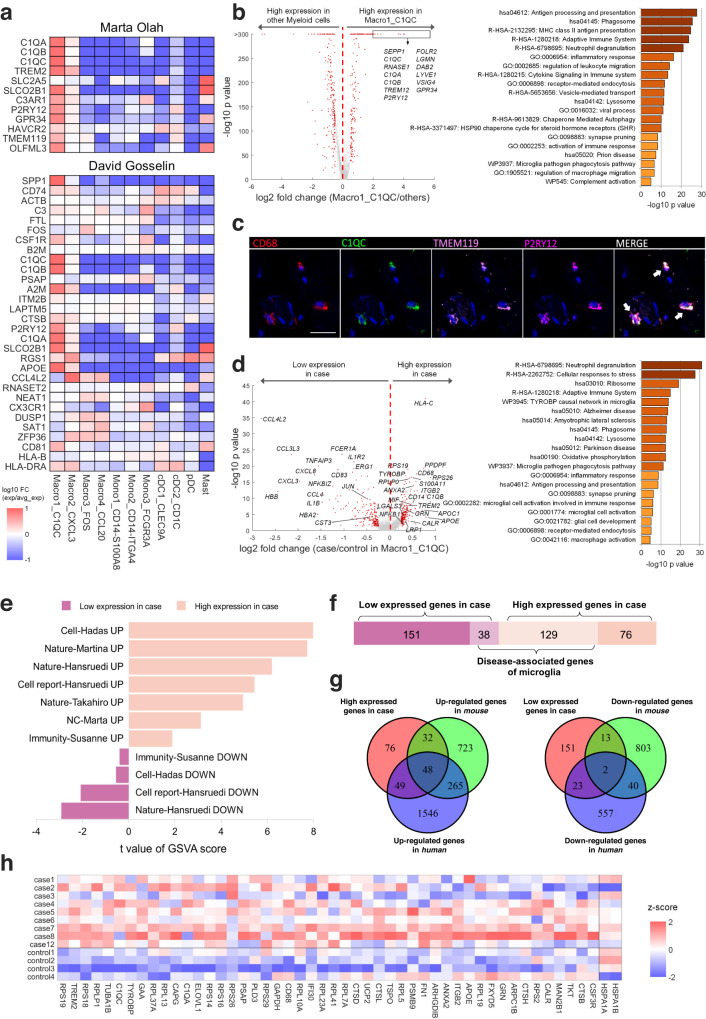
Fig. 3. The C1QC+ macrophages were transcriptionally similar to microglia and exhibited a neurodegenerative dysfunctional phenotype in achalasia.
a Comparison of marker genes of myeloid clusters with previously identified human microglial gene sets reported by Marta Olah and David Gosselin. b Volcano plot (left) and Metascape analysis (right) of marker genes for C1QC+ macrophages compared with other myeloid clusters. c Co-localization of CD68 (red), C1QC (green), TMEM119 (pink) and P2RY12 (magenta) in C1QC+ macrophages of an achalasia patient. Scale bar, 20 μm. Arrow, C1QC+ macrophages. d Volcano plot (left) and Metascape analysis (right) of the DEGs for C1QC+ macrophages between achalasia and controls. e Gene set variation analysis (GSVA) analysis comparing DEGs for C1QC+ macrophages with signatures of neurodegenerative dysfunctional microglia. f, g Comparison of DEGs for C1QC+ macrophages with neurodegenerative dysfunctional genes of microglia by bar chart (f) and Venn diagram (g). h Heatmap showing the specific DEGs for C1QC+ macrophages that overlapped with neurodegenerative dysfunctional genes of microglia. Statistics: Volcano plots (b-left, d-left) were calculated by a two-sided Wilcoxon rank-sum test; Metascape analysis (b-right, d-right) was calculated by Fisher’s exact test. Source data are provided as a Source Data file.