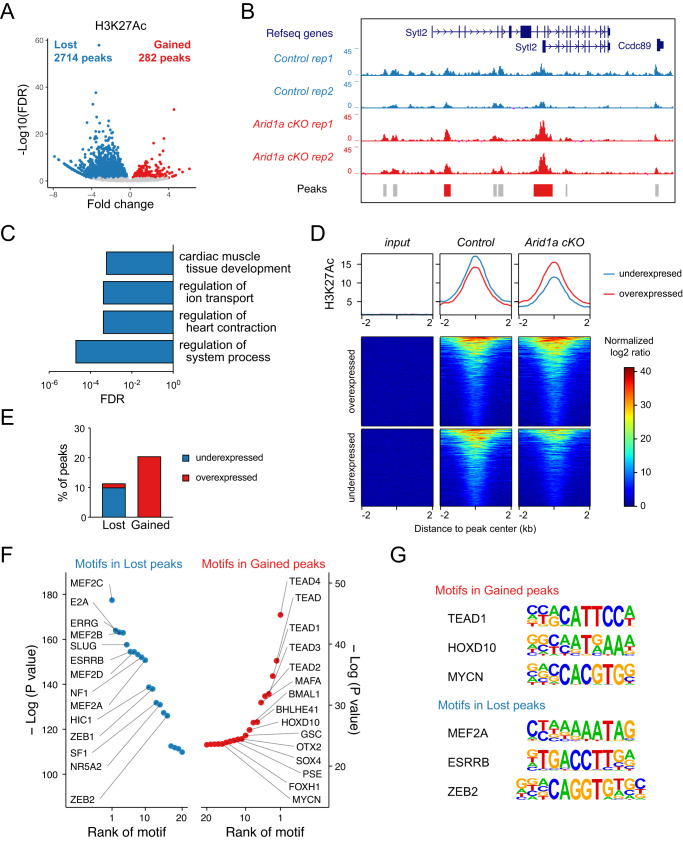
Fig. 5. Loss of Arid1a changes the chromatin landscape in cardiac maturation.
A Volcano plot of H3K27Ac ChIP-Seq on P7 hearts identifies 282 peaks with gained H3K27Ac signal (Gained peaks, red) and 2714 peaks with reduced H3K27Ac levels (Lost peaks, blue) in Arid1a cKO compared to control hearts (Wald test with Benjamini-Hochberg adjusted P-values (false discovery rate (FDR)) < 0.05). B Representative example of H3K27Ac signal within the Sytl2 genomic locus in two replicates (rep) of P7 control (blue) and Arid1a cKO (red) hearts. Peaks are shown below (not changed: gray; peaks with increased H3K27Ac in Arid1a cKO hearts: red). C Gene ontology analysis for biological processes associated with lost peaks. Cardiac muscle development and contraction, and ion handling are enriched. D Average H3K27Ac peak signals (top) and heatmaps for genes overexpressed (middle) or underexpressed (bottom) in Arid1a cKO hearts compared to controls. H3K27Ac in overexpressed genes is increased in Arid1a cKO hearts, whereas H3K27Ac in underexpressed genes is decreased compared to controls. E Intersection of RNA-Seq and H3K27Ac ChIP-Seq indicates that lost peaks are associated with downregulated genes, whereas gained peaks are linked to overexpressed genes. F Motif enrichment analysis shows transcription factor motifs enriched in lost peaks (left, blue) and gained peaks (right, red), ranked by increasing P-value (Hypergeometric Optimization of Motif EnRichment (HOMER) determined P-values of motif enrichment). G Distinct DNA binding motifs of enriched factors in gained peaks and lost peaks. Source data are provided as a Source Data file.