FIG. 4.
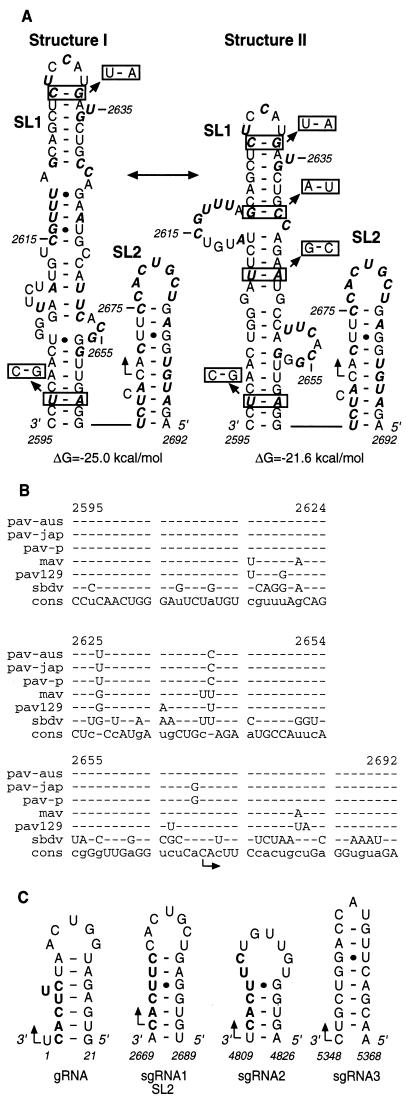
RNA sequence and secondary structure analysis of the sgRNA1 promoter of BYDV. (A) Two secondary structures (I and II), with the calculated free energies predicted with MFOLD (49), contain two stem-loops, SL1 and SL2. Bases in boldface italics differ among Luteovirus members (mostly SbDV [B]). Boxed base pairs indicate covariations that preserve the predicted secondary structure. The right-angled arrow indicates the initiation site (nt 2670). (B) Alignment of RNA sequences in the subgenomic promoter regions of five BYDV strains and SbDV. The BYDV strains are PAV-Australia (also known as PAV-Vic) (pav-aus), PAV-Japan (pav-jap), PAV-Purdue (pav-p), PAV-129 (pav129), and MAV (mav). The bottom row shows consensus sequence (cons). Dashes indicate bases that do not differ from consensus. (C) Computer-predicted stem-loop structures in the genomic and subgenomic sgRNA1 promoter regions of BYDV. The sequence is negative sense; numbering is positive sense. The conserved hexanucleotides at the initiation sites are in boldface.