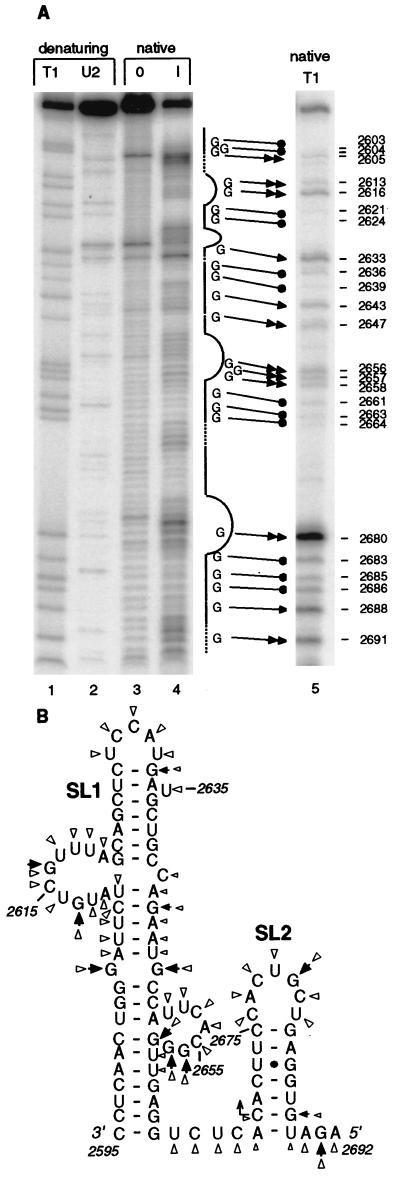
FIG. 5.
Nuclease probing of the sgRNA1 promoter secondary structure. (A) Imidazole and T1 RNase partial digests of 5′-end-labeled transcript of pT7SGP1 containing the sgRNA1 promoter region (negative sense). Gel-purified, end-labeled RNA was incubated in 0 M (0) and 0.8 M (I) imidazole under nondenaturing (native) conditions for 17 h (lanes 3 and 4). The nondenaturing T1 digest was performed with 0.01 U of the enzyme for 5 min at 37°C (lane 5) (see Materials and Methods). Denaturing digests with the T1 (cuts after G) and U2 (cuts A > U) RNases generated markers in lanes 1 and 2. The products were separated on a 6% polyacrylamide gel containing 8 M urea. The straight line beside lane 4 indicates predicted base-paired regions; dashed lines represent predicted single-stranded junctions and ambiguous regions; curved lines show predicted loops and bulges. Double and single arrows represent G’s that were cleaved strongly and weakly, respectively, by T1 nuclease. Filled circles indicate uncut or very weakly cut G’s. (B) Solution structure of the sgRNA1 promoter. Arrowheads represent the T1 analysis data; triangles represent the imidazole digestion data. Larger and smaller symbols indicate strong and weak cuts, respectively.