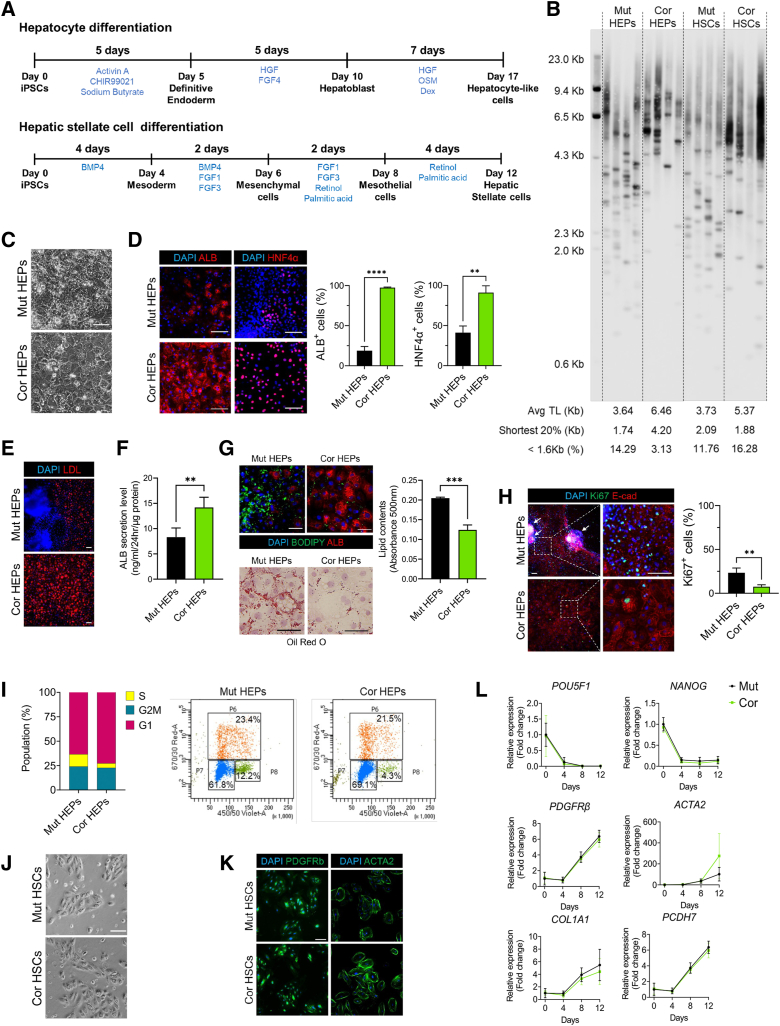
Figure 2.
Differentiation of HEPs and HSCs from isogenic DC iPSCs. (A) Schematic of differentiation scheme for iPSC-derived HEPs and HSCs. (B) Telomere length measurement in HEPs and HSCs by TeSLA. (C) Representative morphologic differences in HEPs on day 17. Scale bar: 100 μm. (D) Immunofluorescence images of ALB and HNF4α in HEPs on day 17. Scale bar: 100 μm. Graphs showing quantification of images (n = 3). (E) Acetylated low-density lipoprotein (LDL) uptake assay in HEPs. Scale bar: 100 μm. (F) ALB secretion by HEPs was analyzed using enzyme-linked immunosorbent assay (n = 4). (G) Lipid accumulation in HEPs analyzed by BODIPY staining and Oil Red O staining. Scale bars: 100 μm. (H) Ki67 and E-cadherin staining in HEPs. White arrows indicate nodule-like structures, quantified at right (n = 3). Scale bar: 100 μm. (I) EdU cell proliferation assays using flow cytometry analysis. (J) Representative morphology of HSCs during the expansion stage (p1). Scale bar: 100 μm. (K) Staining of HSC cultures for stellate cell markers PDGFRβ and ACTA2. Scale bar: 100 μm. (L) qRT-PCR analysis of pluripotency markers and stellate cell marker genes in HSC cultures. For all panels, error bars indicate means ± SD. ∗∗P < .01, ∗∗∗P < .001. Cor, corrected; Mut, mutant; TL, telomere length.